

The Out of East Asia theory of modern human origins supported by recent ancient mtDNA findings
Received date: 2019-03-18
Revised date: 2019-06-16
Online published: 2020-09-10
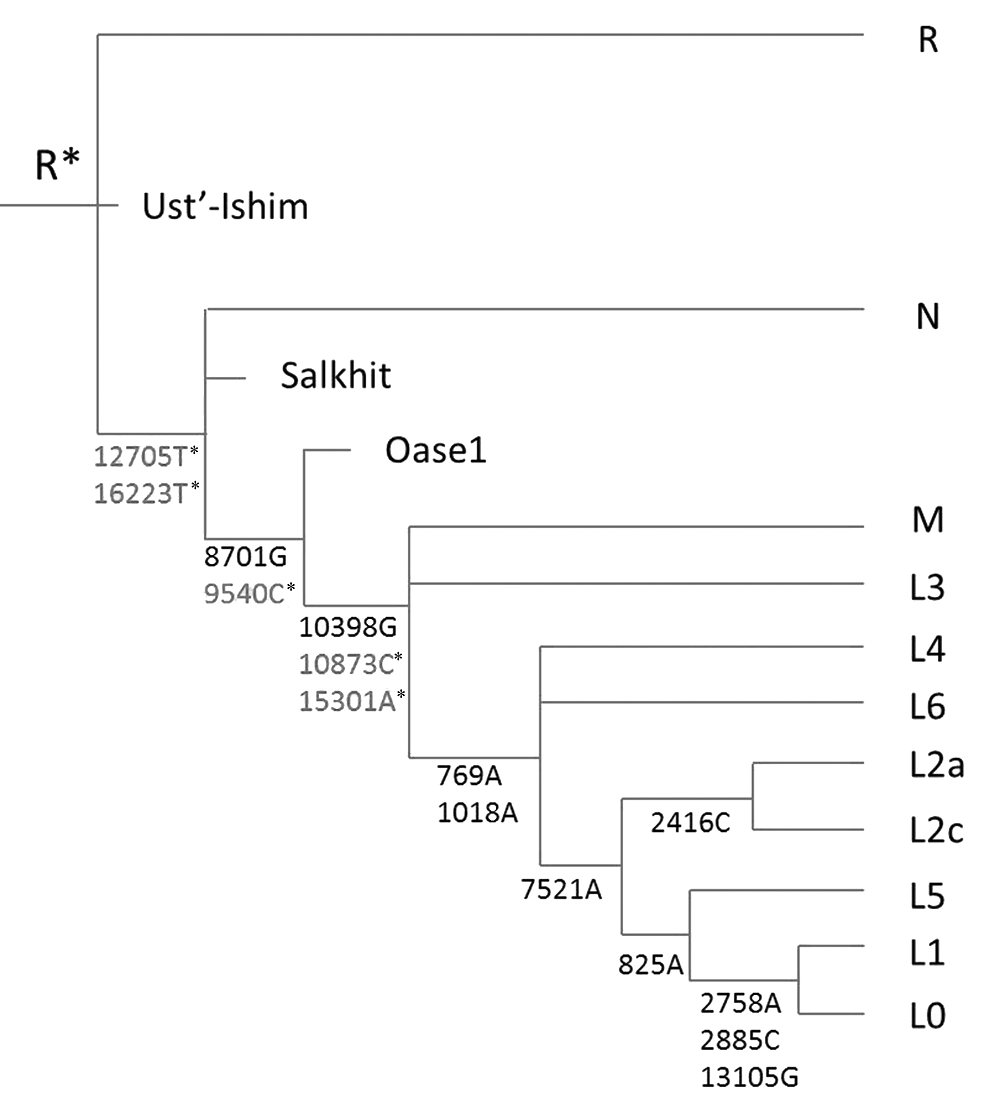
It was in 1983 that scientists constructed the first molecular model of modern human origin based on the mitochondrial DNA (mtDNA) phylogenetic tree, and concluded that modern people originated in Asia. However, in 1987, the Out of Africa model also known as African Eve model was proposed and replaced the original model. But the infinite site assumption and the molecular clock hypothesis on which the African Eve model was based have been widely considered to be unrealistic. In recent years, we have proposed a new molecular evolution theory, namely the maximum genetic diversity(MGD) theory, and used it to reconstruct a new model of human origin, which is basically consistent with the Multiregional model and has the root of modern humans in East Asia. The main difference between the African Eve and our Asia model in the mtDNA tree is the relationship between haplotypes N and R. The African Eve model says that N is the ancestor of R, and our model indicates it is the opposite. In this research, we studied those mtDNA data published from ancient samples, focusing on the relationship between mitochondrial haplogroup N and R. The results show that the three oldest humans (one from 45,000 years ago and the other two about 40,000 years) belong to the haplogroup R. In the human samples from 39,500 to 30,000 years ago, most of them belong to the sub-haplogroup U downstream of the haplogroup R, and only two of them fall into the haplogroup N group(Oase1 is 39,500 years ago, Salkhit is 34,426 years ago). The haplotypes of these two individuals are not part of any prensent N downstream haplotypes and so may be close to the roots of the haplogroup N. These ancient DNA data reveal that the haplogroup R is about 5,000 years older than the haplogroup N, thus confirming the East Asia model and invalidating the African Eve model.
Ye ZHANG , Shi HUANG . The Out of East Asia theory of modern human origins supported by recent ancient mtDNA findings[J]. Acta Anthropologica Sinica, 2019 , 38(04) : 491 -498 . DOI: 10.16359/j.cnki.cn11-1963/q.2019.0068
| [1] | Stringer CB, Andrews P. Genetic and fossil evidence for the origin of modern humans[J]. Science, 1988,239(4845):1263-1268 |
| [2] | Thorne AG, Wolpoff MH. Regional continuity in Australasian Pleistocene hominid evolution[J]. Am J Phys Anthropol, 1981,55:337-349 |
| [3] | Wolpoff MH, Wu XZ, Thorne AG. Modern homo sapiens origins: a general theory of hominid evolution involving the fossil evidence from east Asia[M]. New York: Alan R. Liss, 1984 |
| [4] | Wu X. On the origin of modern humans in China[J]. Quaternary International, 2004,117:131-140 |
| [5] | Yuan D, Lei X, Gui Y, et al. Modern human origins: multiregional evolution of autosomes and East Asia origin of Y and mtDNA[J]. bioRxiv, 2017: doi: https://doi.org/10.1101/106864 |
| [6] | Cann RL, Stoneking AC, Wilson AC. Mitochondrial DNA and human evolution[J]. Nature, 1987,325:31-36 |
| [7] | Green RE, Krause J, Et., et al. A draft sequence of the Neandertal Genome[J]. Science, 2010,328:710-722 |
| [8] | Meyer M, Kircher M, Gansauge MT, et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual[J]. Science, 2012 |
| [9] | Fu Q, Hajdinjak M, Moldovan OT, et al. An early modern human from Romania with a recent Neanderthal ancestor[J]. Nature, 2015,524(7564):216-219 |
| [10] | Fu Q, Li H, Moorjani P, et al. Genome sequence of a 45,000-year-old modern human from western Siberia[J]. Nature, 2014,514(7523):445-449 |
| [11] | Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes[J]. Science, 2014,343(6174):1017-21 |
| [12] | Hublin JJ, Ben-Ncer A, Bailey SE, et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens[J]. Nature, 2017,546(7657):289-292 |
| [13] | White TD, Asfaw B, Degusta D, et al. Pleistocene Homo sapiens from Middle Awash, Ethiopia[J]. Nature, 2003,423:742-747 |
| [14] | Scerri EML, Thomas MG, Manica A, et al. Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter?[J]. Trends Ecol Evol, 2018,33(8):582-594 |
| [15] | Liu W, Martinon-Torres M, Cai YJ, et al. The earliest unequivocally modern humans in southern China[J]. Nature, 2015,526(7575):696-699 |
| [16] | Johnson MJ, Wallace DC, Ferris SD, et al. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns[J]. J Mol Evol, 1983,19(3-4):255-271 |
| [17] | Ayala FJ. Molecular clock mirages[J]. BioEssays, 1999,21(1):71-75 |
| [18] | Ayala FJ. On the virtues and pitfalls of the molecular evolutionary clock[J]. J Hered, 1986,77(4):226-235 |
| [19] | Hu T, Long M, Yuan D, et al. The genetic equidistance result, misreading by the molecular clock and neutral theory and reinterpretation nearly half of a century later[J]. Sci China Life Sci, 2013,56:254-261 |
| [20] | Huang S. The genetic equidistance result of molecular evolution is independent of mutation rates[J]. J. Comp. Sci. Syst. Biol. , 2008,1:92-102 |
| [21] | Huang S. The overlap feature of the genetic equidistance result, a fundamental biological phenomenon overlooked for nearly half of a century[J]. Biological Theory, 2010,5:40-52 |
| [22] | Kumar S. Molecular clocks: four decades of evolution[J]. Nat Rev Genet, 2005,6(8):654-662 |
| [23] | Hahn MW. Toward a selection theory of molecular evolution[J]. Evolution, 2008,62(2):255-265 |
| [24] | Kreitman MW. The neutral theory is dead. Long live the neutral theory[J]. Bioessays, 1996, 18(8): 678-83; discussion 683 |
| [25] | Ohta T, Gillespie JH. Development of Neutral and Nearly Neutral Theories[J]. Theor Popul Biol, 1996,49(2):128-142 |
| [26] | Ballard JW, Kreitman M. Is mitochondrial DNA a strictly neutral marker?[J]. Trends Ecol Evol, 1995,10(12):485-488 |
| [27] | Leffler EM, Bullaughey K, Matute DR, et al. Revisiting an old riddle: what determines genetic diversity levels within species?[J]. PLoS Biol, 2012,10(9):e1001388 |
| [28] | Huang S. New thoughts on an old riddle: What determines genetic diversity within and between species?[J]. Genomics, 2016,108(1):3-10 |
| [29] | Zhu Z, Lu Q, Wang J, et al. Collective effects of common SNPs in foraging decisions in Caenorhabditis elegans and an integrative method of identification of candidate genes[J]. Sci. Rep., 2015: doi: 10.1038/srep16904 |
| [30] | Zhu Z, Yuan D, Luo D, et al. Enrichment of Minor Alleles of Common SNPs and Improved Risk Prediction for Parkinson’s Disease[J]. PLoS ONE, 2015,10(7):e0133421 |
| [31] | Zhu Z, Man X, Xia M, et al. Collective effects of SNPs on transgenerational inheritance in Caenorhabditis elegans and budding yeast[J]. Genomics, 2015,106(1):23-9 |
| [32] | Biswas K, Chakraborty S, Podder S, et al. Insights into the dN/dS ratio heterogeneity between brain specific genes and widely expressed genes in species of different complexity[J]. Genomics, 2016,108(1):11-7 |
| [33] | Zhu Z, Lu Q, Zeng F, et al. Compatibility between mitochondrial and nuclear genomes correlates wtih quantitative trait of lifespan in Caenorhabditis elegans[J]. Sci. Rep., 2015: doi: 10.1038/srep17303 |
| [34] | Huang S. Primate phylogeny: molecular evidence for a pongid clade excluding humans and a prosimian clade containing tarsiers[J]. Sci China Life Sci, 2012,55:709-725 |
| [35] | Huang S. Inverse relationship between genetic diversity and epigenetic complexity[J]. Preprint available at Nature Precedings 2009: doi.org/10.1038/npre.2009.1751.2 |
| [36] | Huang S. Histone methylation and the initiation of cancer, Cancer Epigenetics[M]. New York: CRC Press, 2008 |
| [37] | Margoliash E. Primary structure and evolution of cytochrome c[J]. Proc. Natl. Acad. Sci. , 1963,50:672-679 |
| [38] | Luo D, Huang S. The genetic equidistance phenomenon at the proteomic level[J]. Genomics, 2016,108(1):25-30 |
| [39] | Yuan D, Huang S. On the peopling of the Americas: molecular evidence for the Paleoamerican and the Solutrean models[J]. bioRxiv, 2017, bioRxiv 130989; doi: https://doi.org/10.1101/130989 |
| [40] | Zhang Y, Lei X, Chen H, et al. Ancient DNAs and the Neolithic Chinese super-grandfather Y haplotypes[J]. bioRxiv, 2018, doi: https://doi.org/10.1101/487918 |
| [41] | Meyer M, Fu Q, Aximu-Petri A, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos[J]. Nature, 2014,505(7483):403-406 |
| [42] | Teske P R, Golla T R, Sandoval-Castillo J, et al. Mitochondrial DNA is unsuitable to test for isolation by distance[J]. Sci Rep, 2018,8(1):8448 |
| [43] | Towarnicki SG, Ballard JWO. Mitotype Interacts With Diet to Influence Longevity, Fitness, and Mitochondrial Functions in Adult Female Drosophila[J]. Front Genet, 2018,9:593 |
| [44] | Fu Q, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China[J]. Proc Natl Acad Sci U S A, 2013,110(6):2223-2227 |
| [45] | Benazzi S, Slon V, Talamo S, et al. Archaeology. The makers of the Protoaurignacian and implications for Neandertal extinction[J]. Science, 2015,348(6236):793-796 |
| [46] | Deviese T, Massilani D, Yi S, et al. Compound-specific radiocarbon dating and mitochondrial DNA analysis of the Pleistocene hominin from Salkhit Mongolia[J]. Nat Commun, 2019,10(1):274 |
| [47] | Hershkovitz I, Weber GW, Quam R, et al. The earliest modern humans outside Africa[J]. Science, 2018,359(6374):456-459 |
| [48] | Groucutt H S, Grun R, Zalmout I a S, et al. Homo sapiens in Arabia by 85,000 years ago[J]. Nat Ecol Evol, 2018,2(5):800-809 |
| [49] | Athreya S, Wu X. A multivariate assessment of the Dali hominin cranium from China: Morphological affinities and implications for Pleistocene evolution in East Asia[J]. Am J Phys Anthropol, 2017,164(4):679-701 |
| [50] | Zhao L, Zhang L, Du B, et al. New discovery of human fossils and associated mammal faunas in Bijie Guizhou[J]. Acta Anthropologica Sinica, 2016,35:24-35 |
| [51] | Curnoe D, Ji X, Shajin H, et al. Dental remains from Longtanshan cave 1 (Yunnan, China), and the initial presence of anatomically modern humans in East Asia[J]. Quaternary International, 2016,400:180-186 |
| [52] | Bae CJ, Wang W, Zhao J, et al. Modern human teeth from Late Pleistocene Luna Cave(Guangxi, China)[J]. Quaternary International, 2014,354:169-183 |
| [53] | Li Z Y, Wu XJ, Zhou LP, et al. Late Pleistocene archaic human crania from Xuchang, China[J]. Science, 2017,355(6328):969-972 |
| [54] | Fu Q, Posth C, Hajdinjak M, et al. The genetic history of Ice Age Europe[J]. Nature, 2016,534(7606):200-205 |
| [55] | Posth C, Renaud G, Mittnik A, et al. Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe[J]. Curr Biol, 2016,26(6):827-833 |
/
| 〈 |
|
〉 |