

Osteons and osteocyte lacunae in belanger’s treeshrews (Tupaia belangeri chinensis) - A qualitative image comparative study
Received date: 2020-07-30
Revised date: 2020-09-22
Online published: 2020-11-18
In recent years, treeshrews have gained interest among researchers in the study of human development and disease owed to their phylogenetic closeness to primates. In this comparative study among a mouse, dog, human, baboon, and treeshrews, bone microstructure and morphology were quantitatively analyzed to assess the closeness of treeshrews to humans. In the femurs of three adult male Belanger's treeshrews (Tupaia belangeri chinensis), the osteon structure of the cortical bone was studied using confocal imaging via Fluorescein Iso-Thio-Cyanate sample preparation and staining. Osteocyte lacunae morphology was visualized using acid-etched SEM. Overall, the density and structure of osteon-like formation as well as the morphology of osteocyte lacunae in Belanger's treeshrews bore greater resemblance to mice than humans. These findings indicate that although treeshrews are phylogenetically closer to humans than mice, their bone morphology and functionality are still close to those of mice. This scenario was the first time that osteons and osteocyte lacunae were visualized and characterized in addition to those in a dog and baboon, which enriches our understanding of bone development, adaptation, and evolution in early primates. Future quantitative comparative study is warranted to characterize the micromorphology of bone in treeshrews.
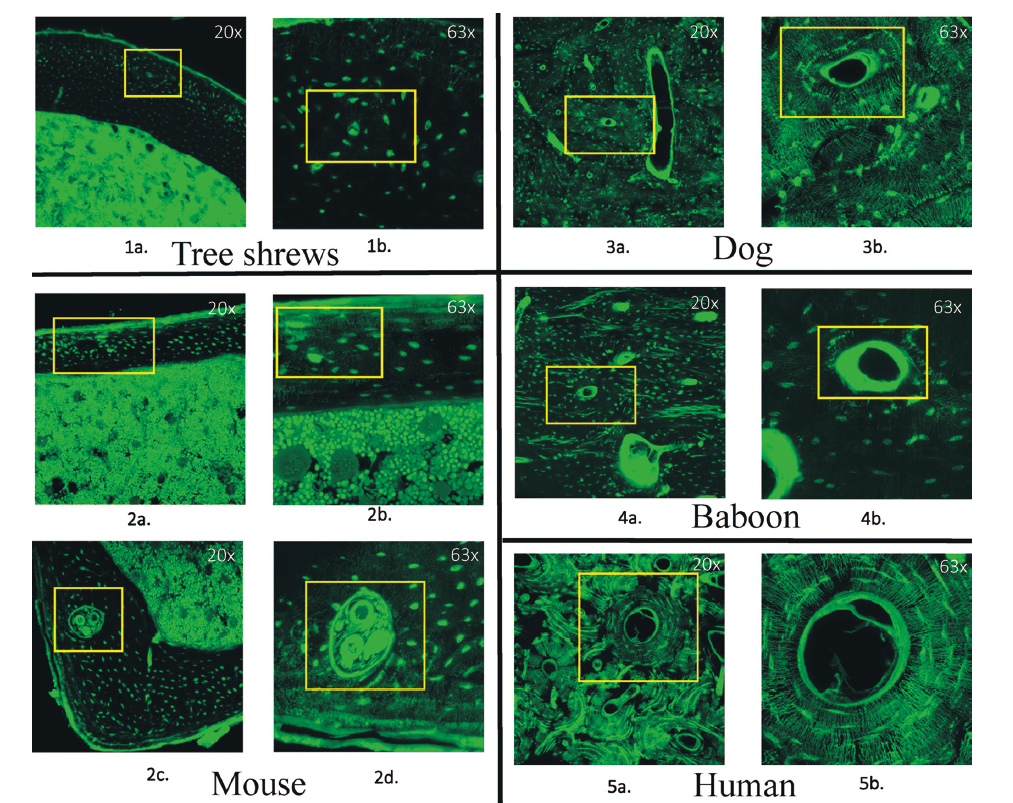
Key words: Bone tissue histology; Cortical bone; Non-human primate model
Lakshmi Pavani MOLLI Vijaya , JAIN Anubhav , Jiangnan FU , Yingjie WU , Q. FENG Jian , Qian WANG . Osteons and osteocyte lacunae in belanger’s treeshrews (Tupaia belangeri chinensis) - A qualitative image comparative study[J]. Acta Anthropologica Sinica, 2020 , 39(04) : 564 -575 . DOI: 10.16359/j.cnki.cn11-1963/q.2020.0048
| [1] | Janecka JE, Miller W, Pringle TH, et al. Molecular and genomic data identify the closest living relative of primates[J]. Science, 2007,318(5851):792-794 |
| [2] | Li Q, Ni X. An Early Oligocene fossil demonstrates treeshrews are slowly evolving “living fossils”[J]. Sci Rep, 2016,6:18627. DOI: 10.1038/srep18627 |
| [3] | Ni X, Qiu Z. Tupaiine treeshrews (Scadentia, Mammalia) from the Yuamnou Lufengpithecus locality of Yunnan, China[J]. Swiss J Palaeontol, 2012,131:51-60 |
| [4] | Zhou X, Sun F, Xu S, et al. The position of treeshrews in the mammalian tree: Comparing multi gene analysis with phylogenomic results leaves monophyly of Euarchonta doubtful[J]. Integr Zool, 2015,10:186-198 |
| [5] | Cao J, Yang EB, Su JJ, et al. The treeshrews: adjuncts and alternatives to primates as models for biomedical research[J]. J Med Primatol, 2003,32(3):123-130 |
| [6] | Yao YG. Creating animal models, why not use the Chinese treeshrew (Tupaia belangeri chinensis)[J]? Zool Res, 2017,38(3):118-126 |
| [7] | Samuels BC, Siegwart JT, Zhan W. et al. A Novel Treeshrew (Tupaia Belangeri) Model of Glaucoma[J]. Invest Opthalmol Vis Sci, 2018,59:3136-3143 |
| [8] | Jiang LP, Shen QS, Yang CP, et al. Establishment of basal cell carcinoma animal model in Chinese tree shrew (Tupaia belangeri chinensis)[J]. Zool Res, 2017,38(4), 180-190 |
| [9] | Ge GZ, Xia HJ, He BL, et al. Generation and characterization of a breast carcinoma model by PyMT overexpression in mammary epithelial cells of treeshrew, an animal close to primates in evolution[J]. Int J Cancer, 2016,138:642-651 |
| [10] | Fan Y, Luo R, Su LY, et al. Does the Genetic Feature of the Chinese Tree Shrew (Tupaia belangeri chinensis) Support its Potential as a Viable Model for Alzheimer’s Disease Research?[J]. J Alzheimers Dis, 2018,61:1015-1028 |
| [11] | Tu Q, Yang D, Zhang X, et al. A novel pancreatic cancer model originated from transformation of acinar cells in adult tree shrew, a primate-like animal[J]. Dis Model Mech, 2019, 12:(4): dmm038703. DOI: 10.1242/dmm.038703 |
| [12] | Tang B, Wu T, Xiao SF, et al. Using Tree Shrews (Tupaia belangeri) as a Novel Animal Model of Liver Transplantation[J]. Curr Med Sci, 2018,38:1069-1074 |
| [13] | Yuan B, Yang C, Xia X, et al. The treeshrews is a promising model for the study of influenza B virus infection[J]. Virol J, 2019,16:77. DOI: 10.1186/s12985-019-1171-3 |
| [14] | Xiao J, Liu R, Chen CS. Tree shrew (Tupaia belangeri) as a novel laboratory disease animal model[J]. Zool Res, 2017,38(3):127-137 |
| [15] | Capulli M, Paone R, Rucci N. Osteoblast and osteocyte: games without frontiers[J]. Arch Biochem Biophys, 2014,561:3-12 |
| [16] | Prideaux M, Findlay DM, Atkins GJ. Osteocytes: The master cells in bone modelling[J]. Curr Opin Pharmacol, 2016,28:24-30 |
| [17] | Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling[J]. Bone, 2008,42:606-615 |
| [18] | Lewis KJ, Frikha-Benayed D, Louie J, et al. Osteocyte calcium signals encode strain magnitude and loading frequency in vivo[J]. Proc Natl Acad Sci USA, 2017,114:11775-11780 |
| [19] | Li X, Zhang Y, Kang H, et al. Sclerostin binds to LRP5/6 and antagonizes canaonical Wnt signalling[J]. J Biol Chem, 2005,280:19883-19887 |
| [20] | Li X, Liu P, Liu W, et al. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation[J]. Nat Genet, 2005,37:945-952 |
| [21] | Nakashima T, Hayashi M, Fukunaga T, et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression[J]. Nat Med, 2011,17(10):1231-1234 |
| [22] | Gluhak-Heinrich J, Pavlin D, Yang W, et al. MEPE expression in osteocytes during orthodontic tooth movement[J]. Arch Oral Biol, 2007,52(7):684-690 |
| [23] | Hadjiagyrou M, Rightmire EP, Ando T, et al. The E11 osteoblastic lineage marker is differentially expressed during fracture healing[J]. Bone, 2001,29(2):149-154 |
| [24] | Toyosowa S, Shintani S, Fujiwara T, et al. Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts[J]. J Bone Miner Res, 2001,16(11):2017-2026 |
| [25] | Feng JQ, Ward LM, Liu S, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism[J]. Nat Genet, 2006,38(11):1310-1315 |
| [26] | Kalajzic I, Matthews BG, Torreggiani E, et al. In vitro and in vivo approaches to study osteocyte biology[J]. Bone, 2013,54(2):296-306 |
| [27] | Kubek DJ, Gattone VH II, Allen MR. Methodological assessment for Acid Etching for visualizing the osteocyte lacunar- canalicular networks using Scanning Electron Microscopy[J]. Microscopy Research and Technique, 2010,73:182-186 |
| [28] | Ren Y, Lin S, Jing Y, et al. A novel way to statistically analyze morphologic changes in Dmp 1 null osteocytes[J]. Connect Tissue Res, 2014,55 Suppl: 129-133 |
| [29] | Bagi CM, Berryman E, Moalli MR. Comparative bone anatomy of commonly used laboratory animals: implications for drug discovery[J]. Comp Med, 2011,61:76-85 |
/
| 〈 |
|
〉 |