

Ancient genomes reveal the complex genetic history of Prehistoric Eurasian modern humans
Received date: 2022-03-04
Revised date: 2022-08-05
Online published: 2023-06-13
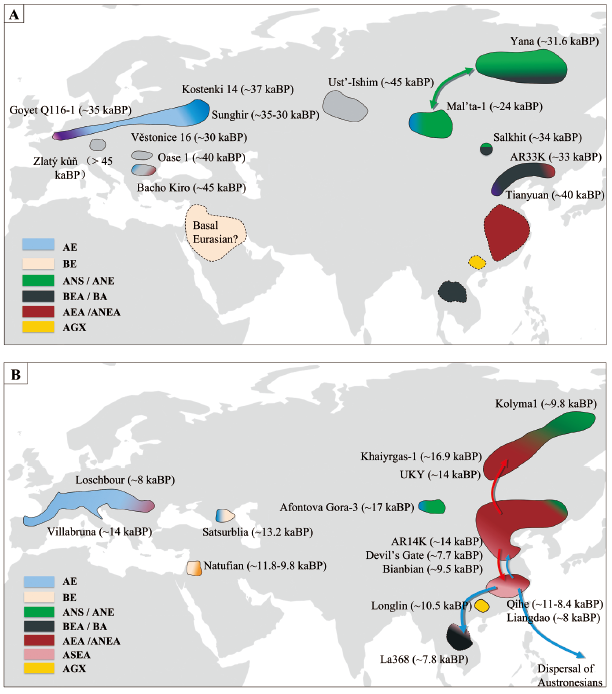
Significant shifts in human populations occurred several times throughout history, as populations dispersed throughout Eurasia about 50 kaBP. During the Last Glacial Maximum (LGM), global temperatures dropped sharply causing environmental deterioration and population turnover in areas. After the LGM, populations increased as the natural environment stabilized and gradually developed into today’s populations. With advancements in ancient DNA extraction and sequencing technology, it is increasingly possible to directly retrieve genome-wide data from prehistoric modern human remains. The rapid emergence of new ancient genomes provides an entirely new direction for studying modern human population structure and evolutionary history. This research on Eurasian populations spanning 45~19 kaBP (pre-LGM) and 19~10 kaBP (post-LGM) summarizes the movement and interaction of prehistoric modern human populations, focusing especially on prehistoric East Eurasia, a region that has been less well-studied genetically. Of at least six distinct populations in Eurasia, three did not contribute substantial ancestry to present-day populations: Ust’-Ishim (≈45 kaBP) from northwestern Siberia; Oase 1 (≈40 kaBP) from Romania; and Zlatý kůň (over 45 kaBP) from Czechia. One population represented by three individuals (4.6~4.3 kaBP, from Bacho Kiro Cave, Bulgaria) seemed to contribute at least a partial genetic component to later some Eurasian populations. One population represented by Tianyuan man (≈40 kaBP, from East Asia) was shown to be more similar to present-day East Asians and Native Americans than to present-day or ancient Europeans. One population represented by Kostenki 14 (≈36 kaBP, from western Siberia) and Goyet Q116-1 (≈35 kaBP, from Belgium) was more closely related to Europeans than to other Eurasians. This work also summarized five representative populations after 40 kaBP and before the end of the LGM. In East Eurasia after the LGM (or since 14 kaBP), population histories played out very differently. For instance, high genetic continuity is observed in the Amur region in the last 14 kaBP, while in the Guangxi region of southern China, an ancient population that lived 10.5 kaBP carried ancestry not represented in any present-day humans. To conclude, comparison of genome-wide ancient DNA from multiple prehistoric humans have illustrated a complex genetic history of prehistoric Eurasian modern humans. In the future, additional ancient genomes will provide more evidence and details to illuminate the complex genetic history of modern humans.
Key words: Eurasia; modern human; Ancient DNA; Population structure; Genetic history
Ming ZHANG , Wanjing PING , Melinda Anna YANG , Qiaomei FU . Ancient genomes reveal the complex genetic history of Prehistoric Eurasian modern humans[J]. Acta Anthropologica Sinica, 2023 , 42(03) : 412 -421 . DOI: 10.16359/j.1000-3193/AAS.2023.0010
| [1] | Nielsen R, Akey JM, Jakobsson M, et al. Tracing the peopling of the world through genomics[J]. Nature, 2017, 541: 302-310 |
| [2] | Sikora M, Pitulko VV, Sousa VC, et al. The population history of northeastern Siberia since the Pleistocene[J]. Nature, 2019, 570: 182-188 |
| [3] | Skoglund P, Mathieson I. Ancient genomics of modern humans: the first decade[J]. Annual Review of Genomics and Human Genetics, 2018, 19: 381-404 |
| [4] | Bradtmoller M, Pastoors A, Weninger B, et al. The repeated replacement model - Rapid climate change and population dynamics in Late Pleistocene Europe[J]. Quaternary International, 2010, 247: 38-49 |
| [5] | Mao XW, Zhang HC, Qiao SY, et al. The deep population history of northern East Asia from the Late Pleistocene to the Holocene[J]. Cell, 2021, 184(e13): 3256-3266 |
| [6] | Green RE, Johannes K, Briggs AW, et al. A draft sequence of the Neandertal Genome[J]. Science, 2010, 328: 710-722 |
| [7] | Fu QM, Hajdinjak M, Moldovan OT, et al. An early modern human from Romania with a recent Neanderthal ancestor[J]. Nature, 2015, 524: 216-219 |
| [8] | Haak W, Lazaridis I, Patterson N, et al. Massive migration from the steppe was a source for Indo-European languages in Europe[J]. Nature, 2015, 522: 207-211 |
| [9] | Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual[J]. Science, 2012, 338: 222-226 |
| [10] | Rasmussen M, Li YR, Lindgreen S, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo[J]. Nature, 2010, 463: 757-762 |
| [11] | Reich D, Green RE, Kircher M, et al. Genetic history of an archaic hominin group from Denisova cave in Siberia[J]. Nature, 2010, 468: 1053-1060 |
| [12] | Liu YC, Mao XW, Johannes K, et al. Insights into human evolution from the first decade of ancient human genomics[J]. Science, 2021, 373: 1479-1484 |
| [13] | Yang MA, Fan XC, Sun B, et al. Ancient DNA indicates human population shifts and admixture in northern and southern China[J]. Science, 2020, 369: 282-288 |
| [14] | Grün R, Stringer C, McDermott F, et al. U-series and ESR analyses of bones and teeth relating to the human burials from Skhul[J]. Journal of Human Evolution, 2005, 49: 316-334 |
| [15] | Liu W, Martinon-Torres M, Cai YJ, et al. The earliest unequivocally modern humans in southern China[J]. Nature, 2015, 526: 696-699 |
| [16] | Mercier N, Valladas H, Bar-Yosef O, et al. Thermoluminescence date for the Mousterian burial site of Es-Skhul, Mt. Carmel[J]. Academic Press, 1993, 20: 169-174 |
| [17] | Fu QM, Li H, Moorjani P, et al. Genome sequence of a 45,000-year-old modern human from western Siberia[J]. Nature, 2014, 514: 445-449 |
| [18] | Prufer K, Posth C, Yu H, et al. A genome sequence from a modern human skull over 45,000 years old from Zlaty kun in Czechia[J]. Nature Ecology & Evolution, 2021, 5: 820-825 |
| [19] | Hajdinjak M, Mafessoni F, Skov L, et al. Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry[J]. Nature, 2021, 592: 253-257 |
| [20] | Fu QM, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan cave, China[J]. Proceedings of the National Academy of Sciences, 2013, 110: 2223-2227 |
| [21] | Shang H, Tong HW, Zhang SQ, et al. An early modern human from Tianyuan cave, Zhoukoudian, China[J]. Proceedings of the National Academy of Sciences, 2007, 104: 6573-6578 |
| [22] | Yang MA, Gao X, Theunert C, et al. 40,000-year-old individual from Asia provides insight into early population structure in Eurasia[J]. Current Biology, 2017, 27: 3202-3208.e9 |
| [23] | Skoglund P, Mallick S, Bortolini MC, et al. Genetic evidence for two founding populations of the Americas[J]. Nature, 2015, 525: 104-108 |
| [24] | Fu QM, Posth C, Hajdinjak M, et al. The genetic history of Ice Age Europe[J]. Nature, 2016, 534: 200-205 |
| [25] | Seguin-Orlando A, Korneliussen TS, Sikora M, et al. Genomic structure in Europeans dating back at least 36,200 years[J]. Science, 2014, 346: 1113-1118 |
| [26] | Sikora M, Seguin-Orlando A, Sousa VC, et al. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers[J]. Science, 2017, 358: 659-662 |
| [27] | Roach JC, Glusman G, Smit AF, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing[J]. Science, 2010, 328: 636-639 |
| [28] | Scally A. The mutation rate in human evolution and demographic inference[J]. Current Opinion in Genetics & Development, 2016, 41: 36-43 |
| [29] | Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution[J]. Nature Reviews Genetics, 2012, 13: 745-753 |
| [30] | Gravel S, Henn BM, Gutenkunst RN, et al. Demographic history and rare allele sharing among human populations[J]. Proceedings of the National Academy of Sciences, 2011, 108: 11983-11988 |
| [31] | Gutenkunst RN, Hernandez RD, Williamson SH, et al. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data[J]. PLOS Genetics, 2009, 5: e1000695 |
| [32] | Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences[J]. Nature Genetics, 2014, 46: 919-925 |
| [33] | Raghavan M, Skoglund P, Graf KE, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans[J]. Nature, 2014, 505: 87-91 |
| [34] | Lipson M, Reich D. A working model of the deep relationships of diverse modern human genetic lineages outside of Africa[J]. Molecular Biology and Evolution, 2017, 34: 889-902 |
| [35] | Massilani D, Skov L, Hajdinjak M, et al. Denisovan ancestry and population history of early East Asians[J]. Science, 2020, 370: 579-583 |
| [36] | Lazaridis I, Patterson N, Mittnik A, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans[J]. Nature, 2014, 513: 409-413 |
| [37] | Lazaridis I, Nadel D, Rollefson G, et al. Genomic insights into the origin of farming in the ancient Near East[J]. Nature, 2016, 536: 419-424 |
| [38] | Wall JD, Yang MA, Flora J, et al. Higher levels of Neanderthal ancestry in East Asians than in Europeans[J]. Genetics, 2013, 194: 199-209 |
| [39] | Vernot B, Akey JM. Complex history of admixture between modern humans and Neandertals[J]. The American Journal of Human Genetics, 2015, 96: 448-453 |
| [40] | Siska V, Jones ER, Jeon S, et al. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago[J]. Science Advances, 2017, 3: e1601877 |
| [41] | Kilinc GM, Kashuba N, Koptekin D, et al. Human population dynamics and Yersinia pestis in ancient northeast Asia[J]. Science Advances, 2021, 7: eabc4587 |
| [42] | Yu H, Spyrou MA, Karapetian M, et al. Paleolithic to Bronze Age Siberians reveal connections with first Americans and across Eurasia[J]. Cell, 2020, 181: 1232-1245.e20 |
| [43] | Zhang M, Fu QM. Human evolutionary history in Eastern Eurasia using insights from ancient DNA[J]. Current Opinion in Genetics & Development, 2020, 62: 78-84 |
| [44] | McColl H, Racimo F, Vinner L, et al. The prehistoric peopling of Southeast Asia[J]. Science, 2018, 361: 88-92 |
| [45] | Damgaard PD, Martiniano R, Kamm J, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia[J]. Science, 2018, 360: eaar7711 |
| [46] | Jones ER, Gonzalez-Fortes G, Connell S, et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians[J]. Nature Communications, 2015, 6: 8912 |
| [47] | Wang TY, Wang W, Xie GM, et al. Human population history at the crossroads of East and Southeast Asia since 11,000 years ago[J]. Cell, 2021, 184(e21): 3829-3841 |
| [48] | Yang MA, Fu QM. Insights into modern human prehistory using ancient genomes[J]. Trends in genetics, 2018, 34: 184-196 |
| [49] | Ning C, Li TJ, Wang K, et al. Ancient genomes from northern China suggest links between subsistence changes and human migration[J]. Nature Communications, 2020, 11: 2700 |
| [50] | Wang CC, Ye HY, Popov AN, et al. Genomic insights into the formation of human populations in East Asia[J]. Nature, 2021, 591: 413-419 |
| [51] | Willerslev E, Meltzer DJ. Peopling of the Americas as inferred from ancient genomics[J]. Nature, 2021, 594: 356-364 |
/
| 〈 |
|
〉 |