| [1] |
肯尼思·F·基普尔. 剑桥世界人类疾病史[M]. 译者:张大庆. 上海: 上海科技教育出版社, 2007: 907-918
|
| [2] |
西里尔·曼戈. 牛津拜占庭史[M]. 译者:陈志强,武鹏 北京: 北京师范大学出版社, 2015: 33-37
|
| [3] |
乔万尼·薄迦丘. 十日谈[M]. 译者:钱鸿嘉,泰和庠,田青. 南京: 译林出版社, 2011: 17-19
|
| [4] |
[东汉]张仲景. 伤寒杂病论[M]. 北京: 中国中医药出版社, 2021: 1-4
|
| [5] |
[南朝·宋范晔]. 后汉书[M]. 北京: 中华书局, 2012: 194-197
|
| [6] |
Arrizabalaga J. The Black Death, 1346-1353: The Complete History[J]. Bulletin of the History of Medicine, 2006, 80(1): 161-163
doi: 10.1353/bhm.2006.0002
URL
|
| [7] |
Allison MJ, Mendoza D, Pezzia A. Documentation of a case of tuberculosis in pre-Columbian America[J]. The American review of respiratory disease, 1973, 107(6): 985-991
|
| [8] |
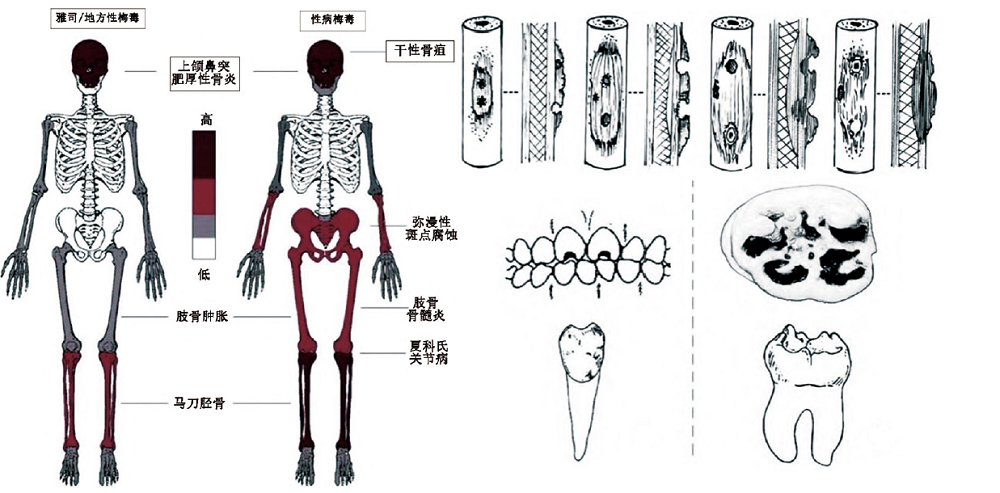
周亚威, 高国帅. 性病梅毒的古病理学研究回顾[J]. 人类学学报, 2022, 41(1): 157-168
|
| [9] |
Bos KI, Schuenemann VJ, Golding GB, et al. A draft genome of Yersinia pestis from victims of the Black Death[J]. Nature, 2011, 478(7370): 506-510
doi: 10.1038/nature10549
URL
|
| [10] |
Fu QM, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China[J]. Proc Natl Acad Sci USA, 2013, 110(6): 2223-2227
doi: 10.1073/pnas.1221359110
URL
|
| [11] |
Burbano HA, Hodges E, Green RE, et al. Targeted investigation of the Neandertal genome by array-based sequence capture[J]. Science, 2010, 328(5979): 723-725
doi: 10.1126/science.1188046
pmid: 20448179
|
| [12] |
Bos KI, Kühnert D, Herbig A, et al. Paleomicrobiology: Diagnosis and Evolution of Ancient Pathogens[J]. Annu Rev Microbiol, 2019, 73: 639-666
doi: 10.1146/annurev-micro-090817-062436
URL
|
| [13] |
Devault AM, McLoughlin K, Jaing C, et al. Ancient pathogen DNA in archaeological samples detected with a Microbial Detection Array[J]. Scientific Reports, 2014, 4: 4245
doi: 10.1038/srep04245
pmid: 24603850
|
| [14] |
Sarkissian CD, Velsko IM, Fotakis AK, et al. Ancient Metagenomic Studies: Considerations for the Wider Scientific Community[J]. mSystems, 2021, 6(6): e01315-21
|
| [15] |
Irving-Pease EK, Muktupavela R, Dannemann M, et al. Quantitative Human Paleogenetics: What can Ancient DNA Tell us About Complex Trait Evolution?[J]. Frontiers in Genetics, 2021, 12: 703541
doi: 10.3389/fgene.2021.703541
URL
|
| [16] |
Duchêne S, Ho SYW, Carmichael AG, et al. The Recovery, Interpretation and Use of Ancient Pathogen Genomes[J]. Current Biology, 2020, 30(19): R1215-R1231
doi: 10.1016/j.cub.2020.08.081
URL
|
| [17] |
武喜艳. 新疆古代致病菌基因组学与进化历史研究[D]. 长春: 吉林大学, 2020: 1-13
|
| [18] |
Tyler AJ, Pe'er I. An Introduction to Whole-Metagenome Shotgun Sequencing Studies[J]. Methods in Molecular Biology, 2021, 2243: 107-122
|
| [19] |
Gaeta R. Ancient DNA and paleogenetics: risks and potentiality[J]. Pathologica, 2021, 113(2): 141-146
doi: 10.32074/1591-951X-146
URL
|
| [20] |
吴斯豪. 新疆塔里木盆地南缘铁器时代人群的基因组学研究[D]. 长春: 吉林大学, 2020: 13-18
|
| [21] |
Firth C, Lipkin WI. The genomics of emerging pathogens[J]. Annual Review of Genomics and Human Genetics, 2013, 14: 281-300
doi: 10.1146/annurev-genom-091212-153446
URL
|
| [22] |
Warinner C, Herbig A, Mann A, et al. A Robust Framework for Microbial Archaeology[J]. Annual Review of Genomics and Human Genetics, 2017, 18: 321-356
doi: 10.1146/annurev-genom-091416-035526
URL
|
| [23] |
Kılınç GM, Kashuba N, Koptekin D, et al. Human population dynamics and Yersinia pestisin ancient northeast Asia[J]. Science Advances, 2021, 7(2): eabc4587
|
| [24] |
Salo WL, Aufderheide AC, Buikstra J, et al. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy[J]. Proc Natl Acad Sci USA, 1994, 91(6): 2091-2094
doi: 10.1073/pnas.91.6.2091
URL
|
| [25] |
Monot M, Honoré N, Garnier T, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae[J]. Nature Genetics, 2009, 41(12): 1282-1289
doi: 10.1038/ng.477
pmid: 19881526
|
| [26] |
Vradenburg JA. The role of treponematoses in the development of prehistoric cultures and the bioarchaeology of proto-urbanism of the central coast of Peru[M]. Columbia: University of Missouri-Columbia, 2001
|
| [27] |
Immel A, Key FM, Szolek A, et al. Analysis of Genomic DNA from Medieval Plague Victims Suggests Long-Term Effect of Yersinia pestis on Human Immunity Genes[J]. Molecular Biology and Evolution, 2021, 38(10): 4059-4076
doi: 10.1093/molbev/msab147
URL
|
| [28] |
Spyrou MA, Bos KI, Herbig A, et al. Ancient pathogen genomics as an emerging tool for infectious disease research[J]. Nature Reviews Genetics, 2019, 20(6): 323-340
doi: 10.1038/s41576-019-0119-1
pmid: 30953039
|
| [29] |
Hansen HB, Damgaard PB, Margaryan A, et al. Comparing Ancient DNA Preservation in Petrous Bone and Tooth Cementum[J]. PLoS One, 2017, 12(1): e0170940
|
| [30] |
Bos KI, Harkins KM, Herbig A, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis[J]. Nature, 2014, 514(7523): 494-497
doi: 10.1038/nature13591
URL
|
| [31] |
Schuenemann VJ, Singh P, Mendum TA, et al. Genome-wide comparison of medieval and modern Mycobacterium leprae[J]. Science, 2013, 341(6142): 179-183
doi: 10.1126/science.1238286
pmid: 23765279
|
| [32] |
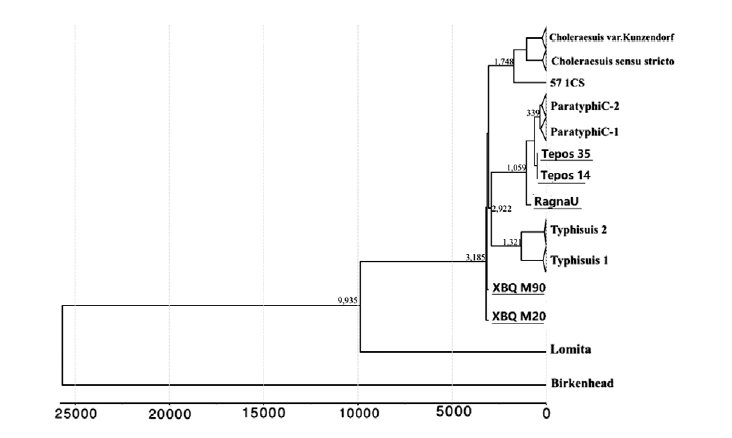
Schuenemann VJ, Lankapalli AK, Barquera R, et al. Historic Treponema pallidum genomes from Colonial Mexico retrieved from archaeological remains[J]. PLoS Neglected Tropical Diseases, 2018, 12(6): e0006447
|
| [33] |
Vågene ÅJ, Herbig A, Campana MG, et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico[J]. Nature Ecology & Evolution, 2018, 2(3): 520-528
|
| [34] |
Marciniak S, Prowse TL, Herring DA, et al. Plasmodium falciparum malaria in 1st-2nd century CE southern Italy[J]. Current Biology, 2016, 26(23): R1220-R1222
doi: 10.1016/j.cub.2016.10.016
URL
|
| [35] |
Maixner F, Kyora BK, Turaev D, et al. The 5300-year-old Helicobacter pylori genome of the Iceman[J]. Science, 2016, 351(6269): 162-165
doi: 10.1126/science.aad2545
pmid: 26744403
|
| [36] |
Duggan AT, Perdomo MF, Piombino-Mascali D, et al. 17th Century Variola Virus Reveals the Recent History of Smallpox[J]. Current Biology, 2016, 26(24): 3407-3412
doi: S0960-9822(16)31324-0
pmid: 27939314
|
| [37] |
Biagini P, Thèves C, Balaresque P, et al. Variola virus in a 300-year-old Siberian mummy[J]. N Engl J Med, 2012, 367(21): 2057-2059
doi: 10.1056/NEJMc1208124
URL
|
| [38] |
Carpenter ML, Buenrostro JD, Valdiosera C, et al. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries[J]. American Journal Of Human Genetics, 2013, 93(5): 852-864
doi: 10.1016/j.ajhg.2013.10.002
pmid: 24568772
|
| [39] |
Key FM, Posth C, Krause J, et al. Mining Metagenomic Data Sets for Ancient DNA: Recommended Protocols for Authentication[J]. Trends in genetics, 2017, 33(8): 508-520
doi: 10.1016/j.tig.2017.05.005
URL
|
| [40] |
Harbeck M, Seifert L, Hänsch S, et al. Yersinia pestis DNA from skeletal remains from the 6(th) century AD reveals insights into Justinianic Plague[J]. PLoS Pathogens, 2013, 9(5): e1003349
|
| [41] |
Giffin K, Lankapalli AK, Sabin S, et al. A treponemal genome from an historic plague victim supports a recent emergence of yaws and its presence in 15th century Europe[J]. Scientific reports, 2020, 10(1): 9499
doi: 10.1038/s41598-020-66012-x
pmid: 32528126
|
| [42] |
Rasmussen S, Allentoft ME, Nielsen K, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago[J]. Cell, 2015, 163(3): 571-582
doi: 10.1016/j.cell.2015.10.009
pmid: 26496604
|
| [43] |
Valtueña AA, Mittnik A, Key FM, et al. The Stone Age Plague and Its Persistence in Eurasia[J]. Current Biology, 2017, 27(23): 3683-3691
doi: 10.1016/j.cub.2017.10.025
URL
|
| [44] |
Luhmann N, Doer D, Chauve C. Comparative scaffolding and gap filling of ancient bacterial genomes applied to two ancient Yersinia pestisgenomes[J]. Microbial Genomics, 2017, 3(9): e000123
|
| [45] |
Song YJ, Wang J, Yang RF, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis[J]. Proc Natl Acad Sci USA, 2013, 110(2): 577-582
doi: 10.1073/pnas.1205750110
URL
|
| [46] |
Wagner DM, Keim PS, Scholz HC, et al. Yersinia pestis and the three plague pandemics--authors' reply[J]. The Lancet Infectious Diseases, 2014, 14(10): 919
|
| [47] |
Damgaard PdB, Marchi N, Rasmussen S, et al. 137 ancient human genomes from across the Eurasian steppes[J]. Nature, 2018, 557(7705): 369-374
doi: 10.1038/s41586-018-0094-2
URL
|
| [48] |
Kirk MD, Pires SM, Black RE, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis[J]. PLoS Medicine, 2015, 12(12): e1001921
|
| [49] |
Zhou ZM, Lundstrøm I, Tran-Dien A, et al. Pan-genome Analysis of Ancient and Modern Salmonella enterica Demonstrates Genomic Stability of the Invasive Para C Lineage for Millennia[J]. Current Biology, 2018, 28(15): 2420-2428
doi: 10.1016/j.cub.2018.05.058
URL
|
| [50] |
Key FM, Posth C, Esquivel-Gomez LR, et al. Emergence of human-adapted Salmonella enterica is linked to the Neolithization process[J]. Nature Ecology & Evolution, 2020, 4(3): 324-333
|
| [51] |
Wu XY, Ning C, Key FM, et al. A 3,000-year-old basal S. enterica lineage from Bronze Age Xinjiang suggests spread along the Proto-Silk Road[J]. PLoS Pathogens, 2021, 17(9): e1009886
|
| [52] |
Spyrou MA, Keller M, Tukhbatova RI, et al. Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes[J]. Nature Communications, 2019, 10(1): 4470
doi: 10.1038/s41467-019-12154-0
pmid: 31578321
|
| [53] |
Susat J, Lübke H, Immel A, et al. A 5,000-year-old hunter-gatherer already plagued by Yersinia pestis[J]. Cell Reports, 2021, 35(13): 109278
doi: 10.1016/j.celrep.2021.109278
URL
|
| [54] |
Stephens JC, Reich DE, Goldstein DB, et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes[J]. American Journal of human genetics, 1998, 62(6): 1507-1515
pmid: 9585595
|
| [55] |
Sabeti PC, Walsh E, Schaffner SF, et al. The case for selection at CCR5-Delta32[J]. PLoS Biology, 2005, 3(11): e378
|
| [56] |
Lindo J, Huerta-Sanchez E, Nakagome S, et al. Demographic and immune-based selection shifts before and after European contact inferred from 50 ancient and modern exomes from the Northwest Coast of North America[J]. BioRxiv 051078
|
| [57] |
Kyora BK, Nutsua M, Boehme L, et al. Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans[J]. Nature Communications, 2018, 9(1): 1569
doi: 10.1038/s41467-018-03857-x
URL
|
| [58] |
Guellil M, Keller M, Dittmar JM, et al. An invasive Haemophilus influenzae serotype b infection in an Anglo-Saxon plague victim[J]. Genome Biology, 2022, 23(1): 22
doi: 10.1186/s13059-021-02580-z
pmid: 35109894
|
| [59] |
Spyrou MA, Tukhbatova RI, Feldman M, et al. Historical Y. pestis Genomes Reveal the European Black Death as the Source of Ancient and Modern Plague Pandemics[J]. Cell Host & Microbe, 2016, 19(6): 874-881
|
| [60] |
Gansauge MT, Meyer M. Selective enrichment of damaged DNA molecules for ancient genome sequencing[J]. Genome research, 2014, 24(9): 1543-1549
doi: 10.1101/gr.174201.114
URL
|
| [61] |
Ginolhac A, Rasmussen M, Gilbert MT, et al. mapDamage: testing for damage patterns in ancient DNA sequences[J]. Bioinformatics, 2011, 27(15): 2153-2155
doi: 10.1093/bioinformatics/btr347
URL
|
| [62] |
Hübler R, Felix MK, Warinner C, et al. HOPS: automated detection and authentication of pathogen DNA in archaeological remains[J]. Genome biology, 2019, 20(1): 280
doi: 10.1186/s13059-019-1903-0
pmid: 31842945
|
| [63] |
Kumar S, Stecher G, Peterson D, et al. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis[J]. Bioinformatics, 2012, 28(20): 2685-2686
doi: 10.1093/bioinformatics/bts507
URL
|
| [64] |
Warinner C, Speller C, Collins MJ. A new era in palaeomicrobiology: prospects for ancient dental calculus as a long-term record of the human oral microbiome[J]. Philos Trans R Soc Lond B Biol Sci, 2015, 370(1660): 20130376
doi: 10.1098/rstb.2013.0376
URL
|