

中国西藏拉托唐古墓地古代居民线粒体全基因组研究
收稿日期: 2019-02-26
修回日期: 2019-05-07
网络出版日期: 2021-02-25
基金资助
国家重点研发计划(2016YFE0203700);国家自然科学基金项目(41672021);国家自然科学基金项目(91731303);国家自然科学基金项目(41630102);中国科学院(QYZDB-SS W-DQC003);中国科学院(XDB26000000);中国科学院(XDB13040000)
A study of the mitochondrial genome of ancient inhabitants from the Latuotanggu cemetery, Tibet, China
Received date: 2019-02-26
Revised date: 2019-05-07
Online published: 2021-02-25
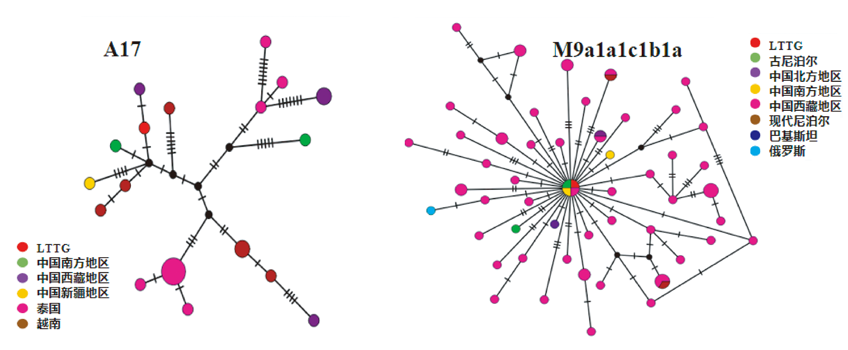
目前青藏高原高海拔地区古DNA研究匮乏。拉托唐古墓地位于青藏高原西南高海拔区域,本文对该墓地出土距今约700年的人骨进行古DNA提取,捕获了高质量线粒体全基因组数据,结合东亚线粒体基因组数据库,运用遗传统计方法开展分析。研究结果表明,距今3000年以内青藏高原西南部人群的遗传历史具有连续性,距今700年左右的拉托唐古墓地居民与距今3150-1250年的古代尼泊尔居民以及现代中国西藏居民母系遗传关系较近,且他们都具有共同的M9a1a1c1b1a单倍群。对M9a1a1c1b1a单倍群的深入研究发现,距今10930-5150年期间青藏高原可能发生了人口扩张事件。以上结果为我们了解古代青藏高原高海拔地区人群遗传历史提供了重要信息。
丁曼雨 , 何伟 , 王恬怡 , 夏格旺堆 , 张明 , 曹鹏 , 刘峰 , 戴情燕 , 付巧妹 . 中国西藏拉托唐古墓地古代居民线粒体全基因组研究[J]. 人类学学报, 2021 , 40(01) : 1 -11 . DOI: 10.16359/j.cnki.cn11-1963/q.2020.0078
With rapid advances in next generation sequencing technologies, we have extracted three ancient DNA from samples from Tibet. With a dataset of present-day East Asians, we tried to reconstruct the history of this region using population genetic methods. Using the mtDNA genome, our study focused on the maternal relationship between ancient Tibetans living in 700BP and present-day populations, which revealed a genetic continuity on Tibetan plateau between the LaTuoTangGu (LTTG) cemetery and present-day Tibetans. This study is the first mtDNA genomic research of high elevation Tibetans.

Key words: Tibet; Ancient DNA; Mitochondrial genome; Latuotanggu; Neolithic
| [1] | Zhang XL, Ha BB, Wang SJ, et al. The earliest human occupation of the high-altitude Tibetan Plateau 40 thousand to 30 thousand years ago[J]. Science, 2018, 362(6418): 1049-1051 |
| [2] | Lu D, Lou H, Kai Y, et al. Ancestral origins and genetic history of Tibetan highlanders[J]. American Journal of Human Genetics, 2016, 99(3): 580-594 |
| [3] | Li J, Zeng W, Zhang Y, et al. Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese[J]. BMC Evolutionary Biology, 2017, 17(1): 239 |
| [4] | Yong-Bin Z, Hong-Jie L, Sheng-Nan L, et al. Ancient DNA evidence supports the contribution of Di-Qiang people to the Han Chinese gene pool[J]. American Journal of Physical Anthropology, 2011, 144(2): 258-268 |
| [5] | Handt O, Krings M, Ward RH, et al. The retrieval of ancient human DNA sequences[J]. American Journal of Human Genetics, 1996, 59(2): 368-76 |
| [6] | Jeong C, Ozga AT, Witonsky DB, et al. Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc[J]. Proceedings of the National Academy of Sciences, 2016, 113(27): 7485 |
| [7] | Duong NT, Macholdt E, Ton ND, et al. Complete human mtDNA genome sequences from Vietnam and the phylogeography of Mainland Southeast Asia[J]. Scientific Reports, 2018, 8(1): 11651. |
| [8] | Lippold S, Xu H, Ko A, et al. Human paternal and maternal demographic histories: Insights from high-resolution Y chromosome and mtDNA sequences[J]. Investigative Genetics, 2014, 5(1): 13. |
| [9] | Zhendong Q, Yajun Y, Longli K, et al. A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum[J]. American Journal of Physical Anthropology, 2010, 143(4): 555-569 |
| [10] | Mian Z, Qing-Peng K, Hua-Wei W, et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau[J]. Proceedings of the National Academy of Sciences, 2009, 106(50): 21230-5 |
| [11] | Peng MS, Palanichamy MG, Yao YG, et al. Inland post-glacial dispersal in East Asia revealed by mitochondrial haplogroup M9a’b[J]. BMC Biology, 2011, 9(1): 2. |
| [12] | Peng MS, Xu W, Song JJ, et al. Mitochondrial genomes uncover the maternal history of the Pamir populations[J]. European Journal of Human Genetics, 2017. |
| [13] | Kang L, Zheng HX, Zhang M, et al. MtDNA analysis reveals enriched pathogenic mutations in Tibetan highlanders[J]. Scientific Reports, 2016, 6(1): 31083 |
| [14] | Li YC, Wang HW, Tian JY, et al. Ancient inland human dispersals from Myanmar into interior East Asia since the Late Pleistocene[J]. Scientific Reports, 2015, 5:9473 |
| [15] | Adimoolam C, Satish K, Jwalapuram S, et al. Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: Dispersal of modern human in South Asian corridor[J]. Plos One, 2009, 4(10): e7447 |
| [16] | Summerer M, Horst J, Erhart G, et al. Large-scale mitochondrial DNA analysis in southeast Asia reveals evolutionary effects of cultural isolation in the multi-ethnic population of Myanmar[J]. BMC Evolutionary Biology, 2014, 14(1): 17 |
| [17] | Wang HW, Li YC, Sun F, et al. Revisiting the role of the Himalayas in peopling Nepal: Insights from mitochondrial genomes[J]. Journal of Human Genetics, 2012, 57(4): 228 |
| [18] | Bhandari S, Zhang X, Cui C, et al. Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region[J]. Scientific Reports, 2015, 5:16249 |
| [19] | Fornarino S, Pala M, Battaglia V, et al. Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): A reservoir of genetic variation[J]. BMC Evolutionary Biology, 2009, 9(1): 154 |
| [20] | Derenko M, Malyarchuk B, Denisova G, et al. Western Eurasian ancestry in modern Siberians based on mitogenomic data[J]. BMC Evolutionary Biology, 2014, 14(1): 217 |
| [21] | Albert Min-Shan K, Chung-Yu C, Qiaomei F, et al. Early Austronesians: into and out of Taiwan[J]. American Journal of Human Genetics, 2014, 94(3): 426-36 |
| [22] | Kutanan W, Kampuansai J, Srikummool M, et al. Complete mitochondrial genomes of Thai and Lao populations indicate an ancient origin of Austroasiatic groups and demic diffusion in the spread of TaifKadai languages[J]. Human Genetics, 2017, 136(1): 85-98 |
| [23] | Dabney J, Knapp M, Glocke I, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(39): 15758-63 |
| [24] | Rohland N, Harney E, Mallick S, et al. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA[J]. Philosophical Transactions of the Royal Society of London, 2015, 370(1660): 20130624 |
| [25] | Jesse D, Matthias M. Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries[J]. Biotechniques, 2012, 52(2): 87-94 |
| [26] | Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform[J]. Nucleic Acids Research, 2012, 40(1): e3 |
| [27] | Fu Q, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(6): 2223 |
| [28] | Gabriel R, Udo S, Janet K. LeeHom: Adaptor trimming and merging for Illumina sequencing reads[J]. Nucleic Acids Research, 2014, 42(18): e141 |
| [29] | Gabriel R, Udo S, Tomislav M, et al. DeML: Robust demultiplexing of Illumina sequences using a likelihood-based approach[J]. Bioinformatics, 2014, 31(5): 770-2 |
| [30] | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-60 |
| [31] | Anita KBT, Dominic P, Sebastian SN, et al. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups[J]. Human Mutation, 2011, 32(1): 25-32 |
| [32] | Oven MV. PhyloTree Build 17: Growing the human mitochondrial DNA tree[J]. Forensic Science International, Genetics Supplement, 2015. |
| [33] | Kutanan W, et al. New insights from Thailand into the maternal genetic history of mainland southeast Asia. Eur J Hum Genet, 2018, 26(6): 898-911 |
| [34] | Edgar RC. Muscle: Multiple sequence alignment with improved accuracy and speed[A]// Proceedings of the Computational Systems Bioinformatics Conference[C], IEEE, 2004. |
| [35] | Edgar RC. Muscle: A multiple sequence alignment method with reduced time and space complexity[J]. BMC Bioinformatics, 2004, 5:113 |
| [36] | Bandelt HJ, Forster P. HLA. Median-joining networks for inferring intraspecific phylogenies[J]. Molecular Biology and Evolution, 1999, 16(1): 37-48 |
| [37] | Leigh JW, Bryant D, Nakagawa S. Popart: Full-feature software for haplotype network construction[J]. Methods in Ecology and Evolution, 2015, 6(9): 1110-1116 |
| [38] | Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10[J]. Virus Evolution, 2018, 4(1). |
| [39] | Excoffier L, Lischer H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows[J]. Molecular Ecology Resources, 2010, 10(3): 564-567 |
| [40] | Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing[J]. Nature Genetics, 2007, 39(12): 1522-1527 |
| [41] | Sawyer S, Renaud G, Viola B, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(51): 15696 |
| [42] | Patterson N, Moorjani P, Luo Y, et al. Ancient admixture in human history[J]. Genetics, 2012, 192(3): 1065 |
| [43] | David P. jModelTest: Phylogenetic model averaging[J]. Molecular Biology & Evolution, 2008, 25(7): 1253-1256 |
| [44] | Jose Manuel S, Diego D, Taboada GL, et al. jmodeltest.org: Selection of nucleotide substitution models on the cloud[J]. Bioinformatics, 2014, 30(9): 1310-1311 |
| [45] | Meyer MC, et al. Permanent human occupation of the central Tibetan Plateau in the early Holocene[J]. Science, 2017, 355(6320): 64-67 |
| [46] | Brantingham PJ, Xing G, Madsen DB, et al. Late occupation of the high-elevation northern Tibetan plateau based on cosmogenic, luminescence, and radiocarbon ages[J]. Geoarchaeology, 2013, 28(5): 413-431 |
| [47] | Chen FH, et al. , Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P[J]. Science, 2015, 347(6219): 248-250 |
| [48] | Lu H. Colonization of the Tibetan plateau, permanent settlement, and the spread of agriculture: Reflection on current debates on the prehistoric archeology of the Tibetan plateau[J]. Archaeological Research in Asia, 2016, 5:12-15 |
| [49] | Qi X, Cui C, Peng Y, et al. Genetic evidence of Paleolithic colonization and Neolithic expansion of modern humans on the Tibetan plateau[J]. Molecular Biology and Evolution, 2013, 30(8): 1761-78 |
/
| 〈 |
|
〉 |