

古DNA揭示中国西北地区史前人类对野生动物资源的利用
收稿日期: 2023-11-07
修回日期: 2023-12-27
网络出版日期: 2025-02-13
基金资助
国家社科基金重大项目“古动物DNA视角下的丝路文化交流研究”(17ZDA221)
Ancient DNA reveals the utilization of wild animal resources by prehistoric humans in Northwest China
Received date: 2023-11-07
Revised date: 2023-12-27
Online published: 2025-02-13
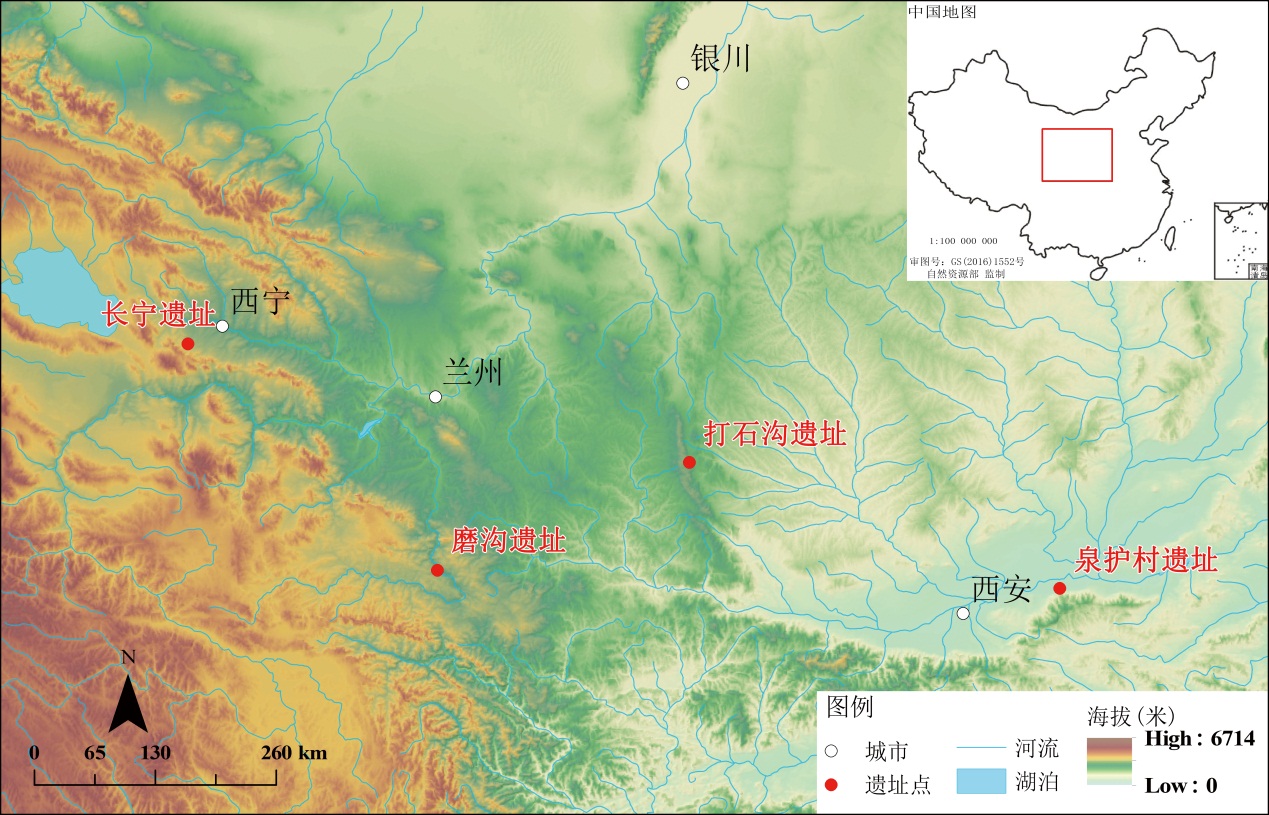
进行古代野生动物遗存的研究,有助于我们了解古代居民的饮食结构、狩猎活动、经济模式等问题,在此基础上应用古DNA技术,则有利于区分家养动物与野生动物。本研究对中国西北地区长宁、磨沟、泉护村、打石沟4个遗址的9例形态学鉴定为“绵羊”或“山羊”的动物样本进行古DNA研究,通过古DNA提取、建库和高通量测序,成功获得了这9例样本的线粒体全基因组序列。根据比对分析、系统发育分析、主成分分析和遗传距离计算,鉴定出这些样本分别为牛科和鹿科动物下的4个不同野生种属,说明古DNA技术可以弥补形态学在鉴定动物遗存方面的不足。结合4个遗址的相关资料,从分子角度证实了中国西北地区史前时期先民在肉食来源、殉葬和骨器原料等方面,普遍存在将野生动物作为动物资源的补充的情况。
宋光捷 , 蔡大伟 , 朱存世 , 胡松梅 , 周静 , 任晓燕 . 古DNA揭示中国西北地区史前人类对野生动物资源的利用[J]. 人类学学报, 2025 , 44(01) : 117 -131 . DOI: 10.16359/j.1000-3193/AAS.2024.0031
Genomic analysis of ancient wild animal remains is of not only great significance for the conservation and utilization of genetic resources of wild animal species but also crucial for helping us understand the diet compositions and hunting activities of ancient human beings as well as their social-economic development patterns. Ancient DNA technology has been widely employed in archaeological research. Among its numerous strengths, its potential to decipher the genetic information carried by biological samples at the molecular level has been widely acknowledged, and many researchers have utilized ancient DNA analysis to distinguish between domestic and wild animals. Moreover, when combined with historical and archaeological evidence, it offers us robust scientific and technological support, enabling us to comprehensively understand ancient human societies, including their origins and evolutionary processes.
In this study, the ancient DNA of nine animal samples, which were excavated from the Changning, Mogou, Quanhucun, and Dashigou sites in Northwest China and morphologically identified as either “sheep” or “goats”, was investigated using ancient DNA technology. Ancient DNA extraction, library construction, and high-throughput sequencing were carried out, and the mitochondrial genome sequences of the nine samples were successfully obtained. Alignment analysis was performed between the genomic sequences of these samples and the 146 mitochondrial genomic sequences of Cervidae and Bovidae (used as reference data). The results of the alignment analysis indicated that these nine samples were identified as belonging to four different wild animal species within the families of Cervidae and Bovidae: the Siberian roe deer (Capreolus pygargus) of the genus Capreolus within the subfamily Odocoileinae; the Przewalski’s gazelle (Procapra przewalskii) and Tibetan gazelle (Procapra picticaudata) of the genus Procapra within the subfamily Antilopinae; and the Sumatran serow (Capricornis sumatraensis) of the genus Capricornis within the subfamily Caprinae.
Two phylogenetic analyses were conducted on the mitochondrial genome data extracted from the Odocoileinae, Caprinae, and Antilopinae subfamilies. Bayesian phylogenetic trees and Maximum Likelihood phylogenetic trees were constructed. It was demonstrated that each of the nine samples clustered with the corresponding species identified by the alignment analysis, which was consistent with the results of the principal component analysis on the same data set, where the nine samples were assigned to the corresponding species identified in the alignment analysis. Genetic distance calculations between individuals based on ancient and modern samples revealed that each of the nine samples was genetically closest to the specific species identified.
All of the above results emphasize that ancient DNA technology can overcome the limitations of morphological methods in the species identification of ancient animals. Considering other wild animal bone remains excavated from the four sites, it can be concluded that the ancient people in prehistoric Northwest China used wild animals as a supplement to domestic animal resources for food, sacrificial offerings, and bone tool manufacture. This study is of great significance as it provides new evidence at the molecular level and corroborates the findings of previous archaeological research on animals in prehistoric times.

| [1] | 蔡大伟, 张乃凡, 赵欣. 中国山羊的起源与扩散研究[J]. 南方文物, 2021, 1: 191-200 |
| [2] | 郭雅琪. 新疆吉仁台沟口遗址出土古代盘羊的线粒体全基因组分析[D].硕士研究生毕业论文, 长春: 吉林大学, 2020, 1-2 |
| [3] | 韩璐. 内蒙古东周时期绵羊和山羊的线粒体DNA研究[D].博士研究生毕业论文, 长春: 吉林大学, 2010, 46-47 |
| [4] | 蔡大伟. 分子考古学导论[M]. 北京: 科学出版社, 2008 |
| [5] | Moore WS. Inferring phylogenies from mtDNA variation: mitochondrial gene trees versus nuclear gene trees[J]. Evolution, 1995, 49: 718-726 |
| [6] | Meadows JRS, Hiendleder S, Kijas JW. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel[J]. Heredity, 2011, 106: 700-706 |
| [7] | E. Dalym G, Torben CR, Nihan DD, et al. Green or white? Morphology, ancient DNA, and the identification of archaeological North American Pacific Coast sturgeon[J]. Journal of Archaeological Science: Reports, 2021, 36: 102887 |
| [8] | 乌日嘎, 臧钰, 孙宏钰. 动物DNA分析技术及其法医学应用[J]. 中国法医学杂志, 2022, 37: 116-120 |
| [9] | 董广辉, 贾鑫, 安邦成, 等. 青海省长宁遗址沉积物元素对晚全新世人类活动和气候变化的响应[J]. 海洋地质与第四纪地质, 2008, 28: 115-119 |
| [10] | 李谅. 青海省长宁遗址的动物资源利用研究[D].硕士研究生毕业论文, 长春: 吉林大学, 2010, 4-6 |
| [11] | 王华, 毛瑞林, 周静. 甘肃临潭磨沟墓地仪式性随葬动物研究[J]. 考古与文物, 2022, 6: 118-125 |
| [12] | 陕西省考古研究院, 渭南市文物旅游局, 华县文物旅游局. 华县泉护村—1997年考古发掘报告[M]. 北京: 文物出版社, 2014 |
| [13] | 杨国宁. 彭阳历史文物[M]. 银川: 宁夏人民教育出版社, 2017 |
| [14] | Yang DY, Eng BW, John S, et al. Improved DNA extraction from ancient bones using silica-based spin columns[J]. American Journal of Physical Anthropology, 1998, 105: 539-543 |
| [15] | Schubert M, Ermini L, Sarkissian CD, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX[J]. Nature Protocols, 2014, 9: 1056-1082 |
| [16] | Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging[J]. BMC Research, 2016, 9: 88 |
| [17] | Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM[CP]. URL: https://doi.org/10.48550/arXiv.1303.3997. Released on: 2013 |
| [18] | Sacco J, Ruplin A, Skonieczny P, et al. Polymorphisms in the canine monoamine oxidase a (MAOA) gene: identification and variation among five broad dog breed groups[J]. Canine Genetics and Epidemiology, 2017, 4: 1 |
| [19] | McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research, 2010, 20: 1297-1303 |
| [20] | Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data[J]. Bioinformatics, 2016, 32: 292-294 |
| [21] | Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of Next Generation Sequencing Data[J]. BMC Bioinformatics, 2014, 15: 356 |
| [22] | Samtools/htslib. lh3/htsbox: My experimental tools on top of htslib[CP]. URL: https://github.com/lh3/htsbox/ |
| [23] | Renaud G, Slon V, Duggan AT, et al. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA[J]. Genome Biology, 2015, 16: 224 |
| [24] | Mpieva/mapping-iterative-assembler: Consensus calling (or “reference assisted assembly”), chiefly of ancient mitochondria[CP]. URL: https://github.com/mpieva/mapping-iterative-assembler/. Released on:2021-03-31 |
| [25] | National Center for Biotechnology Information. BLAST: Basic Local Alignment Search Tool[DB]. URL: https://blastncbi.nlm.nih.gov/ |
| [26] | Seguin-Orlando A, Gamba C, Sarkissian CD. Pros and cons of methylation-based enrichment methods for ancient DNA[J]. Scientific reports, 2015, 5: 11826 |
| [27] | Jónsson H, Ginolhac A, Schubert M, et al. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters[J]. Bioinformatics, 2013, 29: 1682-1684 |
| [28] | Briggs AW, Udo S, Johnson PLF, et al. Patterns of damage in genomic DNA sequences from a Neandertal[J]. Proceedings of the National Academy of Sciences, 2007, 104: 14616-14621 |
| [29] | 朱司褀. 中国北方三个遗址出土马属动物(Equus ovodovi) 的分子考古学研究[D].硕士研究生毕业论文, 长春: 吉林大学, 2020, 46-47 |
| [30] | MikkelSchubert/paleomix. PALEOMIX 1.3.8 Documentation[CP]. URL: https://paleomix.readthedocs.io/en/stable/ |
| [31] | Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Research, 2004, 32: 1792-1797 |
| [32] | Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit[J]. Bioinformatics, 2012, 28: 1166-1167 |
| [33] | Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis[J]. Molecular Biology and Evolution, 2000, 17: 540-552 |
| [34] | Darriba D, Posada D, Kozlov AM, et al. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models[J]. Molecular Biology and Evolution, 2019, 37: 291-294 |
| [35] | Kozlov AM, Darriba D, Flouri T, et al. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference[J]. Bioinformatics, 2019, 35: 4453-4455 |
| [36] | Drummond AJ, Rambaut A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees[J]. BMC Evolutionary Biology, 2007, 7: 214 |
| [37] | BEASTdoc. TreeAnnotator: a program to summarize the information from a sample of trees produced by BEAST onto a single “target” tree[CP]. URL: http://beast.community/treeannotator |
| [38] | Letunic I, Bork P. Interactive tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Research, 2021, 49: W293-W296 |
| [39] | Jombart T. adegenet: a R package for the multivariate analysis of genetic markers[J]. Bioinformatics, 2008, 24: 1403-14-05 |
| [40] | R Core Team. R: A Language and Environment for Statistical Computing[M]. Vienna: R Foundation for Statistical Computing, Austria, 2022 |
| [41] | Wickham H. ggplot2: Elegant Graphics for Data Analysis[M]. New York: Springer-Verlag, 2016 |
| [42] | Tamura K, Stecher G, Kumar S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11[J]. Molecular Biology and Evolution, 2021, 37: 3022-3027 |
| [43] | Kolde R. Pheatmap: Pretty Heatmaps. R package version 1.0.12[CP]. URL: https://CRAN.R-project.org/package=pheatmap. Released on: 2019-01-04 |
| [44] | Castelló JR. Bovids of the World:Antelopes, Gazelles, Cattle, Goats, Sheep, and Relatives[M]. Princeton: Princeton University Press, 2016 |
| [45] | Smith AT. 中国兽类野外手册[M]. 长沙: 湖南教育出版社, 2009 |
| [46] | 蒋志刚. 中国的羊亚科和羚羊亚科动物[J]. 知识就是力量, 2003, 12: 34 |
| [47] | Hewison AJM, Danilkin A. Evidence for separate specific status of European (Capreolus capreolus) and Siberian (C. pygargus) roe deer[J]. Mammalian Biology, 2001, 66: 13-21 |
| [48] | Sheng H. The Deer in China[M]. Shanghai: East China Normal University Press, 1992, 1-251 |
| [49] | 王一如, 乔里斯·彼得斯. 中国西部羊亚科和羚羊亚科颅后骨形态鉴定标准和史前人-羊关系[J]. 南京大学学报(自然科学), 2023, 59: 526-542 |
| [50] | Flad RK, Yuan J, Li S. Zooarcheological evidence for animal domestication in northwest China[J]. Developments in Quaternary Sciences, 2007, 9: 167-203 |
| [51] | 左豪瑞. 中国家羊的动物考古学研究综述和展望[J]. 南方文物, 2017, 1: 155-163 |
| [52] | Taylor WTT, Pruvost M, Posth C, et al. Evidence for early dispersal of domestic sheep into Central Asia[J]. Nature Human Behaviour, 2021, 5: 1169-1179 |
| [53] | 任乐乐. 青藏高原东北部及其周边地区新石器晚期至青铜时代先民利用动物资源的策略研究[D].硕士研究生毕业论文, 兰州: 兰州大学, 2017 |
| [54] | 包曙光. 中国北方地区夏至战国时期的殉牲研究[M]. 北京: 科学出版社, 2021 |
/
| 〈 |
|
〉 |