1 引言
古基因组研究表明,欧亚大陆在史前时期存在多个现代人群体,且这些人群的结构在50 kaBP以来发生了几次重要改变。现代人快速且成功地扩散至整个欧亚大陆乃至于美洲[1⇓-3],可能与这一时期地球气候的几次冷暖更替事件相关[4]。末次盛冰期(约26.5 kaBP开始,一直持续到约19 kaBP)时,全球气温骤降,自然环境极度恶化,一些地区的现代人面临着严峻的生存挑战,有些地区的人群结构可能因此而发生颠覆性的改变,造成冰期后再迁入的人群与之前的人群不存在直接的基因联系[5]。末次盛冰期之后,地球环境进入相对稳定并且适宜人类生存的时期,各地的现代人群体不断发展、迁徙、融合,逐渐形成了现今现代人的群体结构。
近年来,古DNA实验技术与研究方法的高速发展[6⇓⇓-9],极大地推动了人类学的研究。通过古DNA技术直接获取史前材料中的遗传信息,结合材料发现的地点和测年信息,研究者们得以在一个更大的时空框架下开展综合分析。相比于传统的化石形态学研究,古DNA研究对于材料完整度的要求相对较低,并且能够提供更为客观的数据信息。此外,由于每一个个体的古基因组数据都能够在很大程度上反映群体的遗传特征,所以单个个体的古基因组数据能够代表整个群体进行分析,这对于只发现零星人骨遗存的遗址来说,具有更加重要的意义。自2010年,4 kaBP的爱斯基摩人、尼安德特人及丹尼索瓦人的基因组数据发布以来[6,10,11],已经有超过6000例人类古基因组数据发布[12],相关古基因组研究发现和结论已经证实、推翻或修正了古人类学领域许多已有的观点和假说,并提出了许多新的科学认知。如,证实除尼安德特人外还存在另外一种灭绝古人类──丹尼索瓦人[11],发现灭绝古人类(尼安德特人和丹尼索瓦人)与现代人有过基因交流[6,11],推翻了此前认为的现代人与尼安德特人自分离后不再有基因交流的观点;还修正了此前欧亚大陆东部地区的“两层假说”,发现新石器时代早期中国南方福建等沿海地区古人群,实质上并无明显属于“第一层”人群的遗传信息[13]。2017年以前,已发表的古基因组数据主要集中于欧亚大陆西部,科学家们对欧洲、北亚等区域的人群演化问题已展开比较详尽的研究。2017年以来,随着科学家们把关注的重心移向欧亚大陆东部地区,欧亚大陆东部人类古基因组数据开始持续发布,并涌现出了诸多重要的成果,使这一区域人类古基因组研究相对落后的局面已开始逐渐改变。本文重点关注欧亚大陆东部地区史前现代人相关的重要古基因组研究成果,希望为读者梳理、总结欧亚现代人,尤其是欧亚东部现代人在时间与空间上的发展脉络。
2 遗传历史
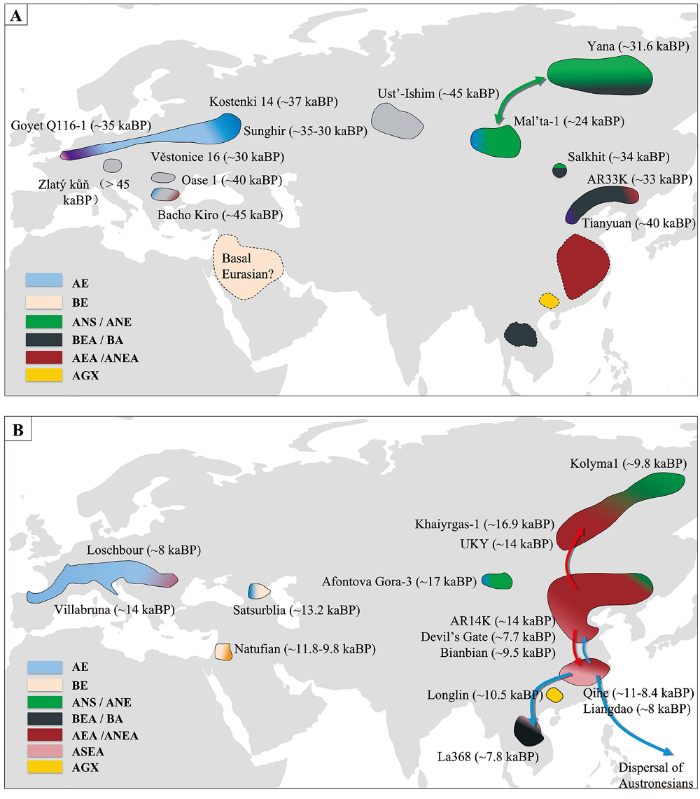
图1
图1
45~10 kaBP欧亚大陆人群示意图
A)阶段1 Period I(45~19 kaBP);B)阶段2 Period II(19~10 kaBP):AE,以Kostenki 14为代表的古欧洲人群相关的遗传成分ancestry related to Ancient Europeans (represent by Kostenki 14);BE,与未经取样的古欧亚人群相关且在现今欧洲人群、部分古代和现今中东人群中可找到的遗传成分ancestry related to an unsampled population known as Basal Eurasians and found in small numbers in ancient and present-day populations of the Near East and in present-day Europeans;ANS,以Yana为代表的古西伯利亚北部人群相关的遗传成分ancestry related to ancient North Siberians (represent by Yana); ANE, ancestry related to ancient North Eurasians (represent by Mal’ta-1 and Afontova Gora-3);ANE,以Mal’ta-1和Afontova Gora-3为代表的古北欧亚人群相关的遗传成分;BEA,以田园洞人和AR33K为代表的基础东亚人相关的遗传成分;BA,以La368为代表的古代东南亚狩猎采集人群相关祖源成分;AEA,东亚古北方人群和东亚古南方人群分离前的亚洲人群;ANEA,以AR14K、扁扁洞、Devil’s Gate等个体为代表的东亚古北方人群;ASEA,以奇和洞、亮岛个体为代表的东亚古南方人群;AGX,以隆林个体为代表的古广西人群。
Fig.1
Schematic of Populations in Eurasia from 45 to 10 kaBP
AE, ; BE, ; ANS, ; BEA, ancestry related to Basal East Asians (represent by Tianyuan and AR33K); BA, ancestry related to Basal Asians (represented by La368); AEA, ancestry related to Ancient East Asians; ANEA, ancestry related to Ancient Northern East Asians (represented by AR14K, Bianbian, Devil’s Gate, etc); ASEA, ancestry related to Ancient Southern East Asians (represented by Qihe and Liangdao); AGX, ancestry related to Ancient Guangxi population (represented by Longlin). The purple color in
2.1 阶段1(45~19 kaBP)
欧亚大陆史前现代人的古基因组研究表明,在约40 kaBP前,欧亚大陆至少存在6个不同的现代人群体(图1),分别为:
同样,Oase 1个体与14 kaBP及更早期的古欧洲人群、现今东亚人群和现今美洲原住民人群均拥有相似数量的等位基因,即拥有相近的亲缘关系。这一结果表明Oase 1个体所代表人群在欧洲无延续至今的直接后裔,是一种对现今欧亚人群无实质遗传贡献的独特人群;而且由于携有6%~9%的尼安德特人遗传成分,Oase 1个体可能代表了与尼安德特人具有密切互动关系的一支欧洲早期现代人群体[7]。
Zlatý kůň个体所代表的人群,同样未给之后的欧洲人和亚洲人贡献基因成分。与同时期的其他现代人个体相似,Zlatý kůň个体携有约3%的尼安德特人成分,但其含有尼安德特人基因片段的平均长度长于Ust-Ishim个体,这说明发生的基因重组次数少,前者的尼安德特人成分混入时间短于后者。Zlatý kůň个体所代表的人群也是最早定居欧亚大陆的人群之一[18]。
2)以发现于保加利亚的46~43 kaBP的Bacho Kiro洞的3个个体为代表的古人群。这一人群相比于晚些时间出现在欧亚大陆西部地区的人群,与东亚和美洲的现代和古代人群关系更近。这表明他们属于一个早前迁入欧洲、但此前没有发现基因证据的群体,而且至少有部分与这一群体相关的基因成分延续到了后期的欧亚大陆人群中[19]。
3)以发现于中国北京约40 kaBP的田园洞人[20⇓-22]为代表的古人群,可称之为“古东亚人”(Basal East Asians, BEA)。这一人群广泛分布在欧亚大陆东部地区,与稍晚一些生存在黑龙江流域的AR33K个体(约33.6 kaBP)属于同一人群。研究发现,田园洞人较之现今欧洲人群和古欧洲人群来说,与现今东亚人群和美洲原住民人群有更近的亲缘关系[22]。相较于其他美洲原住民,某些南美洲原住民人群,特别是苏鲁人(Suruí)和卡利吉亚纳人(Karitiana),显示与大洋洲人、安达曼昂格人和田园洞人均有遗传联系,这表明美洲原住民人群的祖先们可能分批进入美洲,因而亚洲人群对其有不同程度的遗传贡献[22,23]。这些联系也表明,田园洞人的遗传成分至少存在于欧亚东部人群的亚群之中,且以某种形式一直延续到欧洲殖民者进入美洲之前。
4)以发现于西伯利亚西部约36 kaBP的Kostenki 14个体和发现于比利时约35 kaBP的Goyet Q116-1个体[24,25]为代表的古人群,可称之为“古欧洲人”(Ancient Europeans, AE)。“古欧洲人”较之于其他欧亚人群而言,与现今欧洲人群的关系更为相近[24,25],这表明某些与现今欧洲人群有遗传联系的现代人群体至少在35 kaBP已经广泛分布于欧亚大陆西部[24](图1)。发现于俄罗斯约34 kaBP的Sunghir个体与Kostenki 14个体及约34~26 kaBP的捷克Vĕstonice 16个体均有遗传联系,这意味着Kostenki 14所代表人群,至少有一部分被与Vĕstonice 16有亲缘关系的人群所替代[26]。
综合田园洞人、Kostenki 14、Goyet Q116-1等个体的古基因组研究可以发现,在约40 kaBP,遗传学意义上的欧洲人群已在欧洲(比利时)出现,遗传学意义上的亚洲人群也已在亚洲(中国)出现[22]。这也表明欧亚人群的分化(即欧洲人群和亚洲人群的分离)很可能早于40 kaBP,这与依据遗传突变率所推导的这两个人群的分歧时间(40~20 kaBP前至80~40 kaBP)一致[27⇓-29]。然而,虽然田园洞人与Goyet Q116-1个体在地理上相隔万里,但他们之间却存在着遗传联系。此外,他们分别与现今亚洲人群和欧洲人群具有更近的遗传联系,但是他们所共有的等位基因数量比早期亚洲人群和欧洲人群之间以简单分离模式所预期的多[22],这种现象产生的原因可能与亚洲人群和欧洲人群在分离之后仍发生过基因交流相关[30⇓-32]。考虑到Goyet Q116-1个体和田园洞人这两个个体生存时间之早和地理距离之远,Yang等认为当时的东亚人可能并未直接与欧洲人发生基因交流,而可能是跟某一个与这两个个体代表人群均相关的未知人群都发生过基因交流,而这个未知人群可能是从尚未分化的古欧亚人群中的某一亚群演化而来[22]。近期的一项研究发现,保加利亚46~43 kaBP的Bacho Kiro古人群,将Goyet Q116-1个体和田园洞人代表的两个人群联系了起来[19]。
距今4万年以来至末次盛冰期前的现代人群体中有些群体独立存在,还有一些群体与距今4万年以前的古人群相联系,比较具有代表性的有以下5个群体:
2)以发现于蒙古国东北部约34 kaBP的Salkhit个体为代表的古人群。Salkhit个体属于欧亚大陆东西部人群分离后形成的人群,而且有欧亚大陆西部人群的遗传成分。Salkhit个体携带有约75%的与田园洞人相关的祖源成分(BEA相关祖源成分),以及约25%的与Yana人群相关的祖源成分(ANS相关祖源成分)[35]。
3)以发现于中国黑龙江流域约33.6 kaBP的AR33K个体为代表的古人群。AR33K个体与田园洞人有最近的亲缘关系,但是AR33K个体与Goyet Q116-1个体的关系并非像田园洞人与Goyet Q116-1个体一样相近。而且,相比于其他美洲原住民人群,AR33K个体也没有与南美洲的苏鲁人(Surui)拥有更近的关系。另外,AR33K个体中也没有发现与Yana人群相关的祖源成分。这表明东亚北部地区可能在这一时期已经形成复杂的人群结构。田园洞人、AR33K个体及Salkhit个体的古基因组研究共同表明,末次盛冰期前,与田园洞人相关的人群在东亚北部地区广泛分布(分布范围至少覆盖田园洞人代表的华北平原、AR33K代表的中国东北地区和Salkhit代表的蒙古国东北部地区),该人群在蒙古国与Yana相关人群产生了基因混合,而这一时期的黑龙江流域的人群则保持独立特征[5]。
5)以现有遗传数据推测出来的一个未知人群,可称之为古欧亚人群(Basal Eurasians,BE),出现在欧洲人群和亚洲人群分离之前[36]。现今欧洲人群携有这种古欧亚人群的遗传成分,而这种遗传成分的获得主要发生在农业文明进入欧洲以后,因而相较于农业文明以前的古欧洲人群(即14 kaBP及更早期的不携有古欧亚人群(BE)遗传成分的古欧洲人群)而言,现今欧洲人群总体上与其他现代人群的关系较远。Lazaridis等[37]研究发现,古欧亚人群(BE)遗传成分在约34~14 kaBP前的近东人中存在较高比例,在约13 kaBP前高加索地区的狩猎采集人群中也开始发现古欧亚人群(BE)的遗传成分。另外,该人群显示同时具有欧洲人群和近东黎凡特地区的纳图夫人群(Natufians)的遗传特征。欧洲西部的狩猎采集人群与欧洲东部的非洲以外人群(non-African)至少在40 kaBP前就已经分离,而古欧亚人群的分歧时间更早,可能在非洲以外人群与灭绝古人类发生基因混合之前就已经分离[36,37],因此可能不具有或仅具有极低含量的尼安德特人成分[37]。古欧亚人群形成的时间相对较早,存在的时间可能比较长,且持续到更晚的时期,对欧洲和近东地区的现代人产生重要的遗传影响。现今欧洲人群所携有的尼安德特人成分低于现今欧亚地区的其他现代人[9,37⇓-39],很可能是因为古欧亚人群后期进入欧洲,导致欧洲人的尼安德特人成分被稀释。
2.2 阶段2 (19~10 kaBP)
末次盛冰期结束后,欧亚大陆各地区的现代人进入快速发展期。这一时期有些现代人群体与现今现代人群体之间存在一定的遗传连续性,也正是这一时期不同地域古人群的扩增、迁徙、融合等活动,逐渐奠定了现今现代人的遗传结构和分布格局。
在欧亚大陆东北部地区,末次盛冰期前后的人群不连续,但末次盛冰期结束后的人群则存在连续性。发现于中国黑龙江流域19 kaBP的AR19K个体相比于末次盛冰期前的古东亚人(如田园洞人和AR33K)而言,与末次盛冰期后至全新世的人群具有更大的遗传相似性,这表明在末次盛冰期即将结束时,东亚东北部的人群结构已经产生了颠覆性改变[5],这一情况不同于贝加尔湖区域人群在末次盛冰期前后的连续性[24,34]。相比于东亚古南方人群(Ancient Southern East Asians, ASEA;以中国福建约8.4 kaBP的奇和洞个体和台湾海峡约8.3 kaBP的亮岛个体为代表),AR19K个体与东亚古北方人群(Ancient Northern East Asians, ANEA;以中国山东9.5~7.7 kaBP的个体为代表)遗传关系更近。这表明在末次盛冰期结束时,东亚南北方古人群的遗传差异已经形成,而AR19K个体所代表的人群也被认为是最早期的东亚古北方人群(ANEA)[5]。这一人群后期以黑龙江流域的一系列时间连续的个体(AR14K等)、俄罗斯滨海地区(鬼门洞Devil’s Gate)约7.7 kaBP的个体、中国山东9.5~7.7 kaBP的个体(如扁扁洞Bianbian)、贝加尔湖区域约7.1~6.3 kaBP的个体作为代表[2,5,13,40]。自14 kaBP以来,这一人群在中国黑龙江流域具有遗传连续性,现今这一区域的一些少数民族仍然携有这些人群延续下来的遗传成分[40]。
这一时期分布在西伯利亚的古人群显示出不同祖源成分的混合,他们对后期出现的人群产生了重要影响。该人群被称之为古西伯利亚人群(Ancient Paleo-Siberians, APS),以发现于俄罗斯贝加尔湖地区约14 kaBP的UKY个体、远东地区约9.8 kaBP的Kolyma1个体为代表。这一人群以亚洲祖源成分为主,但混有古西伯利亚北部人群(ANS)相关的祖源成分,且Kolyma1代表着在美洲之外与美洲原住民人关系最近的人群[2,42]。在俄罗斯西伯利亚发现的约17 kaBP的Khaiyrgas-1个体所代表的群体与古西伯利亚人群(APS)具有十分相近的遗传关系[41],是该人群的早期代表之一。研究表明古西伯利亚人群(APS)的东亚祖源成分的最近来源便是14 kaBP以来的东亚古北方人群(ANEA)[5],这表明古西伯利亚人群(APS)可能与东亚古北方人群(ANEA)发生过基因混合。而古西伯利亚人群(APS)相关祖源成分也在山东博山(Boshan)、鬼门洞(Devil’s Gate)、贝加尔湖个体中存在,这表明至少有部分遗传成分从古西伯利亚人群(APS)混入到东亚古北方人群(ANEA)中,说明至少在8 kaBP前,古西伯利亚人群(APS)已经与东亚古北方人群(ANEA)有所联系[5,13,43]。
除与现今现代人有遗传联系的古人群外,东亚南部还发现了未对现今现代人产生遗传影响的史前现代人群体。发现于广西隆林(Longlin)约10.5 kaBP的个体所代表的古广西人群(Ancient Guangxi population, AGX),与此前发布的中国南方沿海的东亚古南方人群(ASEA,以此前福建个体为代表)及东南亚狩猎采集人群相关祖源人群(基础亚洲人Basal Asians, BA;以发现于老挝约8 kaBP的La368个体和马来西亚约4.3 kaBP的Ma911个体为代表的Hòabìnhian狩猎采集人群)[44]不同,属于此前未知的东亚现代人群体,对现今现代人未产生遗传影响。这一人群与东亚南北方祖源人群(ASEA和ANEA)具有很深分歧,并且至少持续到6 kaBP。而约9~6 kaBP的广西人群混合有当地祖源成分(AGX)、东亚古南方祖源成分(ASEA)及东南亚狩猎采集人群相关祖源成分(BA)[44],这表明了农业在这一区域兴起之前,人群已经有了广泛的混合,也说明在东亚和东南亚存在的3个独立祖源人群在史前时期互动频繁。
这一时期的欧亚大陆西部人群相比于更古老的欧洲人群,显示出与现今亚洲人群和近东人群更近的亲缘关系。约14 kaBP以来的欧洲中西部个体与一些跨越旧石器和新石器时代的文化个体(如意大利的Villabruna个体)密切相关,且显示出与现今近东人群和东亚人群有遗传联系[22,24,34]。高加索地区约13~10 kaBP的个体(如格鲁吉亚的Satsurblia和Kotias)与欧亚西部个体有较近的亲缘关系,且含有古欧亚人群(BE)的遗传成分。古欧亚人群(BE)的遗传成分在现今欧洲人群和近东人群中均有存在[24,36,37],尤其在近东人群中的含量最高,推算在旧石器时代晚期约12~9.8 kaBP的黎凡特地区,纳图夫人(Natufian)个体中古欧亚人群(BE)遗传成分的含量高达44%;在中石器时代文化约9.1~8.6 kaBP的伊朗个体(如伊朗的Hotu个体)中古欧亚人群(BE)遗传成分含量高达66%[37]。
除以上列举的主要史前现代人群体外,科学家们对全新世以来的更多材料进行了古基因组学研究,发现不同人群之间产生了多次交互影响的事件,对世界现代人群体的分布起到重要的影响。如:在贝加尔湖区域,新石器早期古北欧亚人(ANE)祖源人群,被与现今东亚北部人群相关的祖源人群大量替代[45]。进入全新世之后,东亚南北方人群频繁互动,但整体而言主要有三次大规模的南向基因流动,包括:1)东亚古北方人群相关的遗传成分在东亚南方人群中有所增加;2)具有东亚遗传成分的人群向东南亚扩散,并与基础亚洲人(BA)混合;3)东亚古南方人群祖源成分,通过南岛语系人群向东南亚岛屿和太平洋岛屿扩散[13,43]。人群的互动为现今的东亚大陆人群遗传格局的形成奠定了基础。
3 展望
史前现代人群体的古基因组研究揭示了欧亚大陆丰富的人类历史。对这一时期的现代人展开的古基因组研究,已揭示出欧亚大陆人群丰富多彩的遗传演化历史,极大地提升了我们对史前人群的更替和迁徙历史等相关问题的理解和认识[48]。近年来,欧亚大陆东部的人类古基因组研究快速发展,尤其一些东亚南方样本基因组信息的成功获取,为我们发现和了解更多史前人类群体的遗传历史提供了重要支撑[13,47]。虽然欧亚大陆史前现代人基因混合和人群结构相关的许多问题已得到了答案,但随着研究的深入,新的科学问题也在不断出现,例如,灭绝古人类对不同区域的史前现代人和现今现代人群体的基因贡献程度及影响如何?古基因组是否能发现更多独立的史前现代人群体?史前现代人如何发展、演变、交流,并逐渐形成后期人群及现今人群格局的?
参考文献
Tracing the peopling of the world through genomics
[J].DOI:10.1038/nature21347 [本文引用: 1]
The population history of northeastern Siberia since the Pleistocene
[J].DOI:10.1038/s41586-019-1279-z [本文引用: 5]
Ancient genomics of modern humans: the first decade
[J].
DOI:10.1146/annurev-genom-083117-021749
PMID:29709204
[本文引用: 1]

The first decade of ancient genomics has revolutionized the study of human prehistory and evolution. We review new insights based on prehistoric modern human genomes, including greatly increased resolution of the timing and structure of the out-of-Africa expansion, the diversification of present-day non-African populations, and the earliest expansions of those populations into Eurasia and America. Prehistoric genomes now document population transformations on every inhabited continent-in particular the effect of agricultural expansions in Africa, Europe, and Oceania-and record a history of natural selection that shapes present-day phenotypic diversity. Despite these advances, much remains unknown, in particular about the genomic histories of Asia (the most populous continent) and Africa (the continent that contains the most genetic diversity). Ancient genomes from these and other regions, integrated with a growing understanding of the genomic basis of human phenotypic diversity, will be in focus during the next decade of research in the field.
The repeated replacement model - Rapid climate change and population dynamics in Late Pleistocene Europe
[J].DOI:10.1016/j.quaint.2010.10.015 URL [本文引用: 1]
The deep population history of northern East Asia from the Late Pleistocene to the Holocene
[J].DOI:10.1016/j.cell.2021.04.040 URL [本文引用: 8]
A draft sequence of the Neandertal Genome
[J].
DOI:10.1126/science.1188021
PMID:20448178
[本文引用: 3]

Neandertals, the closest evolutionary relatives of present-day humans, lived in large parts of Europe and western Asia before disappearing 30,000 years ago. We present a draft sequence of the Neandertal genome composed of more than 4 billion nucleotides from three individuals. Comparisons of the Neandertal genome to the genomes of five present-day humans from different parts of the world identify a number of genomic regions that may have been affected by positive selection in ancestral modern humans, including genes involved in metabolism and in cognitive and skeletal development. We show that Neandertals shared more genetic variants with present-day humans in Eurasia than with present-day humans in sub-Saharan Africa, suggesting that gene flow from Neandertals into the ancestors of non-Africans occurred before the divergence of Eurasian groups from each other.
An early modern human from Romania with a recent Neanderthal ancestor
[J].DOI:10.1038/nature14558 [本文引用: 3]
Massive migration from the steppe was a source for Indo-European languages in Europe
[J].DOI:10.1038/nature14317 [本文引用: 2]
A high-coverage genome sequence from an archaic Denisovan individual
[J].
DOI:10.1126/science.1224344
PMID:22936568
[本文引用: 2]

We present a DNA library preparation method that has allowed us to reconstruct a high-coverage (30×) genome sequence of a Denisovan, an extinct relative of Neandertals. The quality of this genome allows a direct estimation of Denisovan heterozygosity indicating that genetic diversity in these archaic hominins was extremely low. It also allows tentative dating of the specimen on the basis of "missing evolution" in its genome, detailed measurements of Denisovan and Neandertal admixture into present-day human populations, and the generation of a near-complete catalog of genetic changes that swept to high frequency in modern humans since their divergence from Denisovans.
Ancient human genome sequence of an extinct Palaeo-Eskimo
[J].DOI:10.1038/nature08835 [本文引用: 1]
Genetic history of an archaic hominin group from Denisova cave in Siberia
[J].DOI:10.1038/nature09710 [本文引用: 3]
Insights into human evolution from the first decade of ancient human genomics
[J].
DOI:10.1126/science.abi8202
URL
[本文引用: 1]

Recent advancements in DNA sequencing technologies and laboratory preparation protocols have rapidly expanded the scope of ancient DNA research over the past decade, both temporally and geographically. Discoveries include interactions between archaic and modern humans as well as modern human population dynamics, including those coinciding with the Last Glacial Maximum and the settlement history of most world regions. This new type of data allows us to examine the deep past of human population dynamics and sharpen the current understanding of our present. The continued development in the ancient DNA field has transformed our understanding of human genetic history and will keep uncovering the further mysteries of our recent evolutionary past.
Ancient DNA indicates human population shifts and admixture in northern and southern China
[J].
DOI:10.1126/science.aba0909
PMID:32409524
[本文引用: 6]

Human genetic history in East Asia is poorly understood. To clarify population relationships, we obtained genome-wide data from 26 ancient individuals from northern and southern East Asia spanning 9500 to 300 years ago. Genetic differentiation in this region was higher in the past than the present, which reflects a major episode of admixture involving northern East Asian ancestry spreading across southern East Asia after the Neolithic, thereby transforming the genetic ancestry of southern China. Mainland southern East Asian and Taiwan Strait island samples from the Neolithic show clear connections with modern and ancient individuals with Austronesian-related ancestry, which supports an origin in southern China for proto-Austronesians. Connections among Neolithic coastal groups from Siberia and Japan to Vietnam indicate that migration and gene flow played an important role in the prehistory of coastal Asia.Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
U-series and ESR analyses of bones and teeth relating to the human burials from Skhul
[J].In order to resolve long-standing issues surrounding the age of the Skhul early modern humans, new analyses have been conducted, including the dating of four well-provenanced fossils by ESR and U-series. If the Skhul burials took place within a relatively short time span, then the best age estimate lies between 100 and 135 ka. This result agrees very well with TL ages obtained from burnt flint of 119+/-18 ka (Mercier et al., 1993). However, we cannot exclude the possibility that the material associated with the Skhul IX burial is older than those of Skhul II and Skhul V. These and other recent age estimates suggest that the three burial sites, Skhul, Qafzeh and Tabun are broadly contemporaneous, falling within the time range of 100 to 130 ka. The presence of early representatives of both early modern humans and Neanderthals in the Levant during Marine Isotope Stage 5 inevitably complicates attempts at segregating these populations by date or archaeological association. Nevertheless, it does appear that the oldest known symbolic burials are those of early modern humans at Skhul and Qafzeh. This supports the view that, despite the associated Middle Palaeolithic technology, elements of modern human behaviour were represented at Skhul and Qafzeh prior to 100 ka.
The earliest unequivocally modern humans in southern China
[J].DOI:10.1038/nature15696 [本文引用: 1]
Thermoluminescence date for the Mousterian burial site of Es-Skhul, Mt. Carmel
[J].
Genome sequence of a 45,000-year-old modern human from western Siberia
[J].DOI:10.1038/nature13810 [本文引用: 3]
A genome sequence from a modern human skull over 45,000 years old from Zlaty kun in Czechia
[J].
Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry
[J].
DOI:10.1038/s41586-021-03335-3
[本文引用: 2]

Modern humans appeared in Europe by at least 45,000 years ago1–5, but the extent of their interactions with Neanderthals, who disappeared by about 40,000 years ago6, and their relationship to the broader expansion of modern humans outside Africa are poorly understood. Here we present genome-wide data from three individuals dated to between 45,930 and 42,580 years ago from Bacho Kiro Cave, Bulgaria1,2. They are the earliest Late Pleistocene modern humans known to have been recovered in Europe so far, and were found in association with an Initial Upper Palaeolithic artefact assemblage. Unlike two previously studied individuals of similar ages from Romania7 and Siberia8 who did not contribute detectably to later populations, these individuals are more closely related to present-day and ancient populations in East Asia and the Americas than to later west Eurasian populations. This indicates that they belonged to a modern human migration into Europe that was not previously known from the genetic record, and provides evidence that there was at least some continuity between the earliest modern humans in Europe and later people in Eurasia. Moreover, we find that all three individuals had Neanderthal ancestors a few generations back in their family history, confirming that the first European modern humans mixed with Neanderthals and suggesting that such mixing could have been common.
DNA analysis of an early modern human from Tianyuan cave, China
[J].
DOI:10.1073/pnas.1221359110
URL
[本文引用: 1]

Hominins with morphology similar to present-day humans appear in the fossil record across Eurasia between 40,000 and 50,000 y ago. The genetic relationships between these early modern humans and present-day human populations have not been established. We have extracted DNA from a 40,000-y-old anatomically modern human from Tianyuan Cave outside Beijing, China. Using a highly scalable hybridization enrichment strategy, we determined the DNA sequences of the mitochondrial genome, the entire nonrepetitive portion of chromosome 21 (∼30 Mbp), and over 3,000 polymorphic sites across the nuclear genome of this individual. The nuclear DNA sequences determined from this early modern human reveal that the Tianyuan individual derived from a population that was ancestral to many present-day Asians and Native Americans but postdated the divergence of Asians from Europeans. They also show that this individual carried proportions of DNA variants derived from archaic humans similar to present-day people in mainland Asia.
An early modern human from Tianyuan cave, Zhoukoudian, China
[J].
DOI:10.1073/pnas.0702169104
URL
[本文引用: 1]

Thirty-four elements of an early modern human (EMH) were found in Tianyuan Cave, Zhoukoudian, China in 2003. Dated to 42,000–39,000 calendrical years before present by using direct accelerator mass spectrometry radiocarbon, the Tianyuan 1 skeleton is among the oldest directly dated EMHs in eastern Eurasia. Morphological comparison shows Tianyuan 1 to have a series of derived modern human characteristics, including a projecting tuber symphyseos, a high anterior symphyseal angle, a broad scapular glenoid fossa, a reduced hamulus, a gluteal buttress, and a pilaster on the femora. Other features of Tianyuan 1 that are more common among EMHs are its modest humeral pectoralis major tuberosities, anteriorly rotated radial tuberosity, reduced radial curvature, and modest talar trochlea. It also lacks several mandibular features common among western Eurasian late archaic humans, including mandibular foramen bridging, mandibular notch asymmetry, and a large superior medial pterygoid tubercle. However, Tianyuan 1 exhibits several late archaic human features, such as its anterior to posterior dental proportions, a large hamulus length, and a broad and rounded distal phalangeal tuberosity. This morphological pattern implies that a simple spread of modern humans from Africa is unlikely.
40,000-year-old individual from Asia provides insight into early population structure in Eurasia
[J].
DOI:S0960-9822(17)31195-8
PMID:29033327
[本文引用: 8]

By at least 45,000 years before present, anatomically modern humans had spread across Eurasia [1-3], but it is not well known how diverse these early populations were and whether they contributed substantially to later people or represent early modern human expansions into Eurasia that left no surviving descendants today. Analyses of genome-wide data from several ancient individuals from Western Eurasia and Siberia have shown that some of these individuals have relationships to present-day Europeans [4, 5] while others did not contribute to present-day Eurasian populations [3, 6]. As contributions from Upper Paleolithic populations in Eastern Eurasia to present-day humans and their relationship to other early Eurasians is not clear, we generated genome-wide data from a 40,000-year-old individual from Tianyuan Cave, China, [1, 7] to study his relationship to ancient and present-day humans. We find that he is more related to present-day and ancient Asians than he is to Europeans, but he shares more alleles with a 35,000-year-old European individual than he shares with other ancient Europeans, indicating that the separation between early Europeans and early Asians was not a single population split. We also find that the Tianyuan individual shares more alleles with some Native American groups in South America than with Native Americans elsewhere, providing further support for population substructure in Asia [8] and suggesting that this persisted from 40,000 years ago until the colonization of the Americas. Our study of the Tianyuan individual highlights the complex migration and subdivision of early human populations in Eurasia.Copyright © 2017 Elsevier Ltd. All rights reserved.
Genetic evidence for two founding populations of the Americas
[J].DOI:10.1038/nature14895 [本文引用: 1]
The genetic history of Ice Age Europe
[J].DOI:10.1038/nature17993 [本文引用: 7]
Genomic structure in Europeans dating back at least 36,200 years
[J].
DOI:10.1126/science.aaa0114
PMID:25378462
[本文引用: 2]

The origin of contemporary Europeans remains contentious. We obtained a genome sequence from Kostenki 14 in European Russia dating from 38,700 to 36,200 years ago, one of the oldest fossils of anatomically modern humans from Europe. We find that Kostenki 14 shares a close ancestry with the 24,000-year-old Mal'ta boy from central Siberia, European Mesolithic hunter-gatherers, some contemporary western Siberians, and many Europeans, but not eastern Asians. Additionally, the Kostenki 14 genome shows evidence of shared ancestry with a population basal to all Eurasians that also relates to later European Neolithic farmers. We find that Kostenki 14 contains more Neandertal DNA that is contained in longer tracts than present Europeans. Our findings reveal the timing of divergence of western Eurasians and East Asians to be more than 36,200 years ago and that European genomic structure today dates back to the Upper Paleolithic and derives from a metapopulation that at times stretched from Europe to central Asia. Copyright © 2014, American Association for the Advancement of Science.
Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers
[J].
DOI:10.1126/science.aao1807
PMID:28982795
[本文引用: 1]

Present-day hunter-gatherers (HGs) live in multilevel social groups essential to sustain a population structure characterized by limited levels of within-band relatedness and inbreeding. When these wider social networks evolved among HGs is unknown. To investigate whether the contemporary HG strategy was already present in the Upper Paleolithic, we used complete genome sequences from Sunghir, a site dated to ~34,000 years before the present, containing multiple anatomically modern human individuals. We show that individuals at Sunghir derive from a population of small effective size, with limited kinship and levels of inbreeding similar to HG populations. Our findings suggest that Upper Paleolithic social organization was similar to that of living HGs, with limited relatedness within residential groups embedded in a larger mating network.Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Analysis of genetic inheritance in a family quartet by whole-genome sequencing
[J].
DOI:10.1126/science.1186802
PMID:20220176
[本文引用: 1]

We analyzed the whole-genome sequences of a family of four, consisting of two siblings and their parents. Family-based sequencing allowed us to delineate recombination sites precisely, identify 70% of the sequencing errors (resulting in > 99.999% accuracy), and identify very rare single-nucleotide polymorphisms. We also directly estimated a human intergeneration mutation rate of approximately 1.1 x 10(-8) per position per haploid genome. Both offspring in this family have two recessive disorders: Miller syndrome, for which the gene was concurrently identified, and primary ciliary dyskinesia, for which causative genes have been previously identified. Family-based genome analysis enabled us to narrow the candidate genes for both of these Mendelian disorders to only four. Our results demonstrate the value of complete genome sequencing in families.
The mutation rate in human evolution and demographic inference
[J].
Revising the human mutation rate: implications for understanding human evolution
[J].
DOI:10.1038/nrg3295
PMID:22965354
[本文引用: 1]

It is now possible to make direct measurements of the mutation rate in modern humans using next-generation sequencing. These measurements reveal a value that is approximately half of that previously derived from fossil calibration, and this has implications for our understanding of demographic events in human evolution and other aspects of population genetics. Here, we discuss the implications of a lower-than-expected mutation rate in relation to the timescale of human evolution.
Demographic history and rare allele sharing among human populations
[J].
DOI:10.1073/pnas.1019276108
URL
[本文引用: 1]

High-throughput sequencing technology enables population-level surveys of human genomic variation. Here, we examine the joint allele frequency distributions across continental human populations and present an approach for combining complementary aspects of whole-genome, low-coverage data and targeted high-coverage data. We apply this approach to data generated by the pilot phase of the Thousand Genomes Project, including whole-genome 2–4× coverage data for 179 samples from HapMap European, Asian, and African panels as well as high-coverage target sequencing of the exons of 800 genes from 697 individuals in seven populations. We use the site frequency spectra obtained from these data to infer demographic parameters for an Out-of-Africa model for populations of African, European, and Asian descent and to predict, by a jackknife-based approach, the amount of genetic diversity that will be discovered as sample sizes are increased. We predict that the number of discovered nonsynonymous coding variants will reach 100,000 in each population after ∼1,000 sequenced chromosomes per population, whereas ∼2,500 chromosomes will be needed for the same number of synonymous variants. Beyond this point, the number of segregating sites in the European and Asian panel populations is expected to overcome that of the African panel because of faster recent population growth. Overall, we find that the majority of human genomic variable sites are rare and exhibit little sharing among diverged populations. Our results emphasize that replication of disease association for specific rare genetic variants across diverged populations must overcome both reduced statistical power because of rarity and higher population divergence.
Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data
[J].DOI:10.1371/journal.pgen.1000695 URL [本文引用: 1]
Inferring human population size and separation history from multiple genome sequences
[J].
DOI:10.1038/ng.3015
PMID:24952747
[本文引用: 1]

The availability of complete human genome sequences from populations across the world has given rise to new population genetic inference methods that explicitly model ancestral relationships under recombination and mutation. So far, application of these methods to evolutionary history more recent than 20,000-30,000 years ago and to population separations has been limited. Here we present a new method that overcomes these shortcomings. The multiple sequentially Markovian coalescent (MSMC) analyzes the observed pattern of mutations in multiple individuals, focusing on the first coalescence between any two individuals. Results from applying MSMC to genome sequences from nine populations across the world suggest that the genetic separation of non-African ancestors from African Yoruban ancestors started long before 50,000 years ago and give information about human population history as recent as 2,000 years ago, including the bottleneck in the peopling of the Americas and separations within Africa, East Asia and Europe.
Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans
[J].DOI:10.1038/nature12736 [本文引用: 2]
A working model of the deep relationships of diverse modern human genetic lineages outside of Africa
[J].
DOI:10.1093/molbev/msw293
PMID:28074030
[本文引用: 3]

A major topic of interest in human prehistory is how the large-scale genetic structure of modern populations outside of Africa was established. Demographic models have been developed that capture the relationships among small numbers of populations or within particular geographical regions, but constructing a phylogenetic tree with gene flow events for a wide diversity of non-Africans remains a difficult problem. Here, we report a model that provides a good statistical fit to allele-frequency correlation patterns among East Asians, Australasians, Native Americans, and ancient western and northern Eurasians, together with archaic human groups. The model features a primary eastern/western bifurcation dating to at least 45,000 years ago, with Australasians nested inside the eastern clade, and a parsimonious set of admixture events. While our results still represent a simplified picture, they provide a useful summary of deep Eurasian population history that can serve as a null model for future studies and a baseline for further discoveries.© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Denisovan ancestry and population history of early East Asians
[J].
DOI:10.1126/science.abc1166
PMID:33122380
[本文引用: 2]

We present analyses of the genome of a ~34,000-year-old hominin skull cap discovered in the Salkhit Valley in northeastern Mongolia. We show that this individual was a female member of a modern human population that, following the split between East and West Eurasians, experienced substantial gene flow from West Eurasians. Both she and a 40,000-year-old individual from Tianyuan outside Beijing carried genomic segments of Denisovan ancestry. These segments derive from the same Denisovan admixture event(s) that contributed to present-day mainland Asians but are distinct from the Denisovan DNA segments in present-day Papuans and Aboriginal Australians.Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Ancient human genomes suggest three ancestral populations for present-day Europeans
[J].DOI:10.1038/nature13673 [本文引用: 4]
Genomic insights into the origin of farming in the ancient Near East
[J].DOI:10.1038/nature19310 [本文引用: 6]
Higher levels of Neanderthal ancestry in East Asians than in Europeans
[J].
DOI:10.1534/genetics.112.148213
PMID:23410836
[本文引用: 1]

Neanderthals were a group of archaic hominins that occupied most of Europe and parts of Western Asia from ∼30,000 to 300,000 years ago (KYA). They coexisted with modern humans during part of this time. Previous genetic analyses that compared a draft sequence of the Neanderthal genome with genomes of several modern humans concluded that Neanderthals made a small (1-4%) contribution to the gene pools of all non-African populations. This observation was consistent with a single episode of admixture from Neanderthals into the ancestors of all non-Africans when the two groups coexisted in the Middle East 50-80 KYA. We examined the relationship between Neanderthals and modern humans in greater detail by applying two complementary methods to the published draft Neanderthal genome and an expanded set of high-coverage modern human genome sequences. We find that, consistent with the recent finding of Meyer et al. (2012), Neanderthals contributed more DNA to modern East Asians than to modern Europeans. Furthermore we find that the Maasai of East Africa have a small but significant fraction of Neanderthal DNA. Because our analysis is of several genomic samples from each modern human population considered, we are able to document the extent of variation in Neanderthal ancestry within and among populations. Our results combined with those previously published show that a more complex model of admixture between Neanderthals and modern humans is necessary to account for the different levels of Neanderthal ancestry among human populations. In particular, at least some Neanderthal-modern human admixture must postdate the separation of the ancestors of modern European and modern East Asian populations.
Complex history of admixture between modern humans and Neandertals
[J].DOI:10.1016/j.ajhg.2015.01.006 URL [本文引用: 1]
Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago
[J].
DOI:10.1126/sciadv.1601877
URL
[本文引用: 2]

Early Neolithic (~7700-year-old) genetic data from the Russian Far East implies a high level of genetic continuity in this region.
Human population dynamics and Yersinia pestis in ancient northeast Asia
[J].
DOI:10.1126/sciadv.abc4587
URL
[本文引用: 1]

\n Ancient genomes reveal a complex demographic picture since the post-LGM in northeast Asia and report the presence of\n Y. pestis\n.\n
Paleolithic to Bronze Age Siberians reveal connections with first Americans and across Eurasia
[J].
DOI:S0092-8674(20)30502-X
PMID:32437661
[本文引用: 1]

Modern humans have inhabited the Lake Baikal region since the Upper Paleolithic, though the precise history of its peoples over this long time span is still largely unknown. Here, we report genome-wide data from 19 Upper Paleolithic to Early Bronze Age individuals from this Siberian region. An Upper Paleolithic genome shows a direct link with the First Americans by sharing the admixed ancestry that gave rise to all non-Arctic Native Americans. We also demonstrate the formation of Early Neolithic and Bronze Age Baikal populations as the result of prolonged admixture throughout the eighth to sixth millennium BP. Moreover, we detect genetic interactions with western Eurasian steppe populations and reconstruct Yersinia pestis genomes from two Early Bronze Age individuals without western Eurasian ancestry. Overall, our study demonstrates the most deeply divergent connection between Upper Paleolithic Siberians and the First Americans and reveals human and pathogen mobility across Eurasia during the Bronze Age.Copyright © 2020 Elsevier Inc. All rights reserved.
Human evolutionary history in Eastern Eurasia using insights from ancient DNA
[J].
The prehistoric peopling of Southeast Asia
[J].
DOI:10.1126/science.aat3628
PMID:29976827
[本文引用: 2]

The human occupation history of Southeast Asia (SEA) remains heavily debated. Current evidence suggests that SEA was occupied by Hòabìnhian hunter-gatherers until ~4000 years ago, when farming economies developed and expanded, restricting foraging groups to remote habitats. Some argue that agricultural development was indigenous; others favor the "two-layer" hypothesis that posits a southward expansion of farmers giving rise to present-day Southeast Asian genetic diversity. By sequencing 26 ancient human genomes (25 from SEA, 1 Japanese Jōmon), we show that neither interpretation fits the complexity of Southeast Asian history: Both Hòabìnhian hunter-gatherers and East Asian farmers contributed to current Southeast Asian diversity, with further migrations affecting island SEA and Vietnam. Our results help resolve one of the long-standing controversies in Southeast Asian prehistory.Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
The first horse herders and the impact of early Bronze Age steppe expansions into Asia
[J].
DOI:10.1126/science.aar7711
URL
[本文引用: 1]

\n The Eurasian steppes reach from the Ukraine in Europe to Mongolia and China. Over the past 5000 years, these flat grasslands were thought to be the route for the ebb and flow of migrant humans, their horses, and their languages. de Barros Damgaard\n et al.\n probed whole-genome sequences from the remains of 74 individuals found across this region. Although there is evidence for migration into Europe from the steppes, the details of human movements are complex and involve independent acquisitions of horse cultures. Furthermore, it appears that the Indo-European Hittite language derived from Anatolia, not the steppes. The steppe people seem not to have penetrated South Asia. Genetic evidence indicates an independent history involving western Eurasian admixture into ancient South Asian peoples.\n
Upper Palaeolithic genomes reveal deep roots of modern Eurasians
[J].
DOI:10.1038/ncomms9912
PMID:26567969

Jones, Eppie R.; Martiniano, Rui; McLaughlin, Russell L.; Cassidy, Lara M.; Gamba, Cristina; Pinhasi, Ron; Bradley, Daniel G. Univ Dublin Trinity Coll, Smurfit Inst Genet, Dublin 2, Ireland. Gonzalez-Fortes, Gloria; Hofreiter, Michael Univ Potsdam, Inst Biochem & Biol, Dept Math & Nat Sci, D-14476 Potsdam, Germany. Gonzalez-Fortes, Gloria Univ Ferrara, Dept Biol & Evolut, I-44100 Ferrara, Italy. Connell, Sarah; Gamba, Cristina; Pinhasi, Ron Univ Coll Dublin, Sch Archaeol, Dublin 4, Ireland. Connell, Sarah; Gamba, Cristina; Pinhasi, Ron Univ Coll Dublin, Earth Inst, Dublin 4, Ireland. Siska, Veronika; Eriksson, Anders; Llorente, Marcos Gallego; Manica, Andrea Univ Cambridge, Dept Zool, Cambridge CB2 3EJ, England. Eriksson, Anders King Abdullah Univ Sci & Technol, Div Biol & Environm Sci & Engn, Integrat Syst Biol Lab, Thuwal 239556900, Saudi Arabia. Gamba, Cristina Univ Copenhagen, Nat Hist Museum Denmark, Ctr GeoGenet, DK-1350 Copenhagen, Denmark. Meshveliani, Tengiz; Jakeli, Nino; Lordkipanidze, David Georgian Natl Museum, GE-0105 Tbilisi, Rep of Georgia. Bar-Yosef, Ofer Harvard Univ, Peabody Museum, Dept Anthropol, Cambridge, MA 02138 USA. Mueller, Werner Univ Neuchatel, Lab Archeozool, CH-2000 Neuchatel, Switzerland. Mueller, Werner LATENIUM, Off Patrimoine & Archeol Neuchatel, Sect Archeol, CH-2068 Hauterive, Switzerland. Belfer-Cohen, Anna Hebrew Univ Jerusalem, Inst Archaeol, IL-91905 Jerusalem, Israel. Matskevich, Zinovi Israel Antiqu Author, IL-91004 Jerusalem, Israel. Higham, Thomas F. G. Univ Oxford, Oxford Radiocarbon Accelerator Unit, Res Lab Archaeol & Hist Art, Oxford OX1 3QY, England. Currat, Mathias Univ Geneva, Anthropol Unit, Dept Genet & Evolut, Lab Anthropol Genet & Peopling Hist AGP, CH-1227 Geneva, Switzerland.
Human population history at the crossroads of East and Southeast Asia since 11,000 years ago
[J].DOI:10.1016/j.cell.2021.05.018 URL [本文引用: 2]
Insights into modern human prehistory using ancient genomes
[J].
DOI:S0168-9525(17)30210-X
PMID:29395378
[本文引用: 1]

The genetic relationship of past modern humans to today's populations and each other was largely unknown until recently, when advances in ancient DNA sequencing allowed for unprecedented analysis of the genomes of these early people. These ancient genomes reveal new insights into human prehistory not always observed studying present-day populations, including greater details on the genetic diversity, population structure, and gene flow that characterized past human populations, particularly in early Eurasia, as well as increased insight on the relationship between archaic and modern humans. Here, we review genetic studies on ∼45000- to 7500-year-old individuals associated with mainly preagricultural cultures found in Eurasia, the Americas, and Africa.Copyright © 2017 Elsevier Ltd. All rights reserved.
Ancient genomes from northern China suggest links between subsistence changes and human migration
[J].
DOI:10.1038/s41467-020-16557-2
PMID:32483115
[本文引用: 1]

Northern China harbored the world's earliest complex societies based on millet farming, in two major centers in the Yellow (YR) and West Liao (WLR) River basins. Until now, their genetic histories have remained largely unknown. Here we present 55 ancient genomes dating to 7500-1700 BP from the YR, WLR, and Amur River (AR) regions. Contrary to the genetic stability in the AR, the YR and WLR genetic profiles substantially changed over time. The YR populations show a monotonic increase over time in their genetic affinity with present-day southern Chinese and Southeast Asians. In the WLR, intensification of farming in the Late Neolithic is correlated with increased YR affinity while the inclusion of a pastoral economy in the Bronze Age was correlated with increased AR affinity. Our results suggest a link between changes in subsistence strategy and human migration, and fuel the debate about archaeolinguistic signatures of past human migration.
Genomic insights into the formation of human populations in East Asia
[J].DOI:10.1038/s41586-021-03336-2 [本文引用: 1]
Peopling of the Americas as inferred from ancient genomics
[J].DOI:10.1038/s41586-021-03499-y [本文引用: 1]